Team:Freiburg/Results
From 2011.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
==<span style="color:red;">red light receptor</span>== | ==<span style="color:red;">red light receptor</span>== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
In August we decided to drop the red light receptor system.<br/> | In August we decided to drop the red light receptor system.<br/> | ||
The main reason beeing Jakob having to leave the team to start studying in Strasbourg.<br/> | The main reason beeing Jakob having to leave the team to start studying in Strasbourg.<br/> | ||
So the FreiGEM team shrunk to five people.<br/> | So the FreiGEM team shrunk to five people.<br/> | ||
| - | |||
Still we did submit one part for this system which is [http://partsregistry.org/Part:BBa_K608151 BBa_K608151] | Still we did submit one part for this system which is [http://partsregistry.org/Part:BBa_K608151 BBa_K608151] | ||
| Line 34: | Line 26: | ||
We talked to some of the iGEM teams like TU Munich also working with this system<br/> | We talked to some of the iGEM teams like TU Munich also working with this system<br/> | ||
and we are sure that the red light receptor is not lost.<br/> | and we are sure that the red light receptor is not lost.<br/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==<span style="color:blue;">blue light receptor</span>== | ==<span style="color:blue;">blue light receptor</span>== | ||
| Line 97: | Line 76: | ||
Please follow the links for more information. | Please follow the links for more information. | ||
| + | ==<span style="color:grey;">Precipitator</span>== | ||
| + | [http://partsregistry.org/Part:BBa_K608404 BBa_K608404] | ||
| + | IPTG-inducible Promoter with plastic binding domain-tagged GFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608406 BBa_K608406] | ||
| + | '''Precipitator | ||
| + | ''' | ||
| + | |||
| + | The Precipitator is a new artificially designed LRR protein. It is meant as protein that binds Nickel ions with Histidines grouped on its surface. The bound Nickel can then precipitate His-tagged proteins. In our Lab in a Cell it should function as an adaptor between the plastic surface of pipettes and the His-tagged protein. Please look at our detailed description of the design layout in our modeling section. The sequence was synthesised and cloned into the iGEm vector. The submitted sequence was fully confirmed by sequencing. | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608407 BBa_K608407] | ||
| + | '''Precipitator fused with GST tag''' | ||
| + | |||
| + | The Precipitator was fused with the C-terminus of our GST tag in order to extract and further test the construct for its Nickel binding affinity. We successfully cloned it together using the Gibson Assembly and then subsequently pasted it into the iGEM vector. The submitted sequence was partially confirmed by sequencing. | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608408 BBa_K608408] | ||
| + | '''GST-tag''' | ||
| + | |||
| + | The GST-tag was PCR amplified from a pGEX vector with overhang primers including the iGEM restriction sites and then pasted into the iGEM vector. To verify the functionality of the construct we cloned it before a GFP sequence and expressed it with an IPTG inducible vector. Results see partsregistry page. The submitted sequence was partially confirmed by sequencing. | ||
| + | |||
| + | ==<span style="color:green;">Green light receptor</span>== | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608101 BBa_K608101] | ||
| + | '''CcaR''', green light response regulator | ||
| + | |||
| + | The green light response regulator CcaR was amplified via PCR from<br/> | ||
| + | the whole ''Synechocystis sp. PCC 6803'' genome and inserted into the pSB1C3 vector. | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608102 BBa_K608102] | ||
| + | CcaS, green light receptor | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608151 BBa_K608151] | ||
| + | translational unit of pcyA | ||
| + | |||
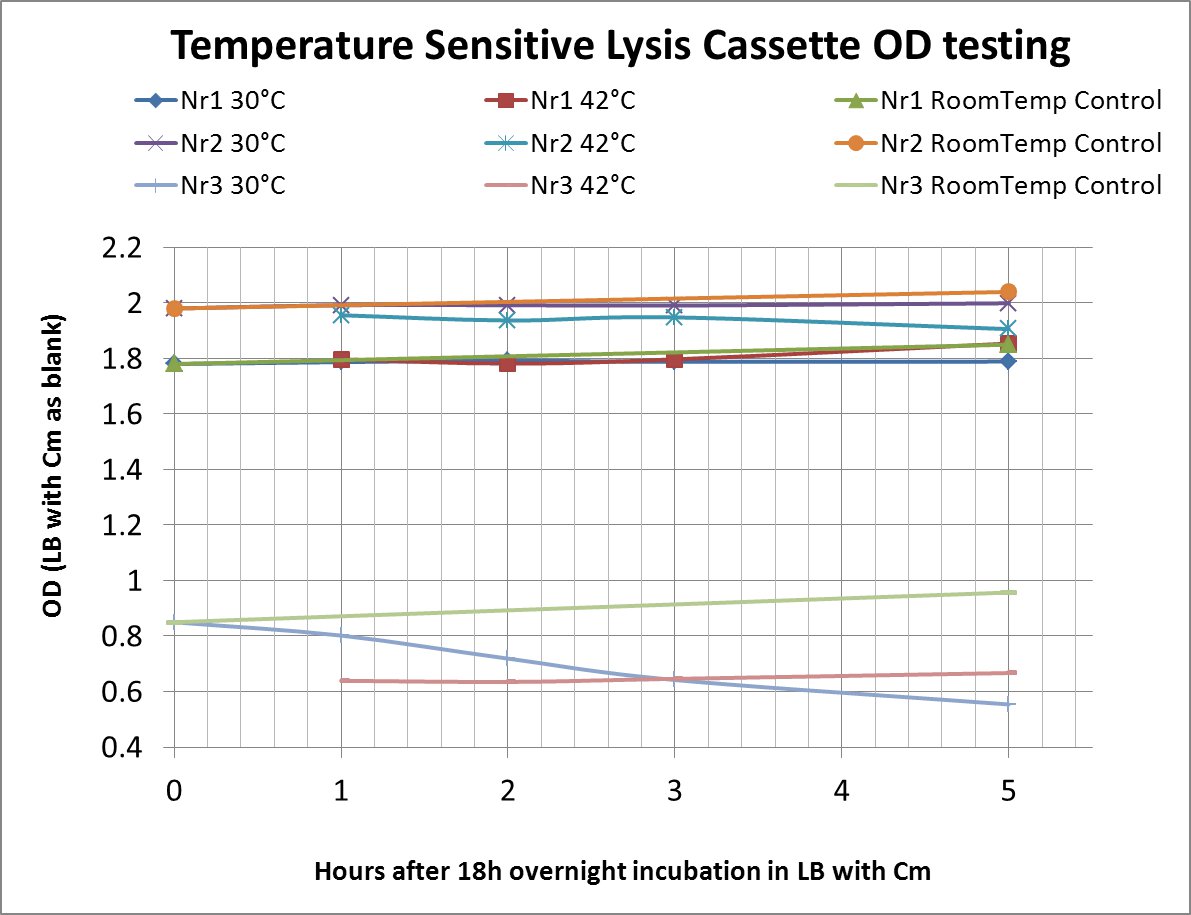
| + | ==<span style="color:orange;">Lysis cassette</span>== | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608351 BBa_K608351] | ||
| + | (correct) temperature sensitive promoter | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608352 BBa_K608352] | ||
| + | Bacteriophage Lysis Cassette with RBS | ||
| + | |||
| + | ==<span style="color:black;">commons</span>== | ||
| + | |||
| + | At the beginning we had to assemble promoters and ribosome binding sites of different "strengths" (ie consensus identities) in order to ensure proper and controlled expression of our parts.<br/> | ||
| + | <br/> | ||
| + | The cloning procedure was time-consuming (circa two weeks), so we decided to<br/> | ||
| + | send our constructs to the registry. Herewith we hope to help others save some time.<br/> | ||
| + | We called these constructs ''PR'' to abbreviate "Promoter and RBS" | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608002 BBa_K608002] | ||
| + | strong Promoter and strong RBS | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608003 BBa_K608003] | ||
| + | strong Promoter , medium RBS | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608004 BBa_K608004] | ||
| + | strong Promoter , weak RBS | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608005 BBa_K608005] | ||
| + | medium Promoter , strong RBS | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608006 BBa_K608006] | ||
| + | medium Promoter , medium RBS | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608007 BBa_K608007] | ||
| + | medium Promoter , weak RBS | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | To quantify the strength of our PR-constructs we cloned GFP and RFP behind it<br/> | ||
| + | and quantified the protein output.<br/> | ||
| + | |||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608008 BBa_K608008] | ||
| + | constitutive strong promoter with medium RBS and GFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608009 BBa_K608009] | ||
| + | Strong promoter with weak RBS and GFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608010 BBa_K608010] | ||
| + | Medium promoter with strong RBS and GFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608011 BBa_K608011] | ||
| + | Medium promoter with medium RBS and GFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608012 BBa_K608012] | ||
| + | Medium promoter with weak RBS and GFP | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608013 BBa_K608013] | ||
| + | strong Promoter with strong RBS and RFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608014 BBa_K608014 ] | ||
| + | PR with RFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608015 BBa_K608015] | ||
| + | PR with RFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608016 BBa_K608016] | ||
| + | PR with RFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608017 BBa_K608017] | ||
| + | PR with RFP | ||
| + | |||
| + | [http://partsregistry.org/Part:BBa_K608018 BBa_K608018] | ||
| + | PR with RFP | ||
{{:Team:Freiburg/Templates/footer}} | {{:Team:Freiburg/Templates/footer}} | ||
Revision as of 00:45, 22 September 2011
 "
"
 Contact
Contact