Team:EPF-Lausanne/Tools/Gibson assembly
From 2011.igem.org
| Line 31: | Line 31: | ||
# '''Negative control:''' For the negative control, make a Gibson reaction on just the plasmid backbone DNA and transform it. This will show you how much of the backbone closes itself up in your actual Gibson reaction. | # '''Negative control:''' For the negative control, make a Gibson reaction on just the plasmid backbone DNA and transform it. This will show you how much of the backbone closes itself up in your actual Gibson reaction. | ||
| - | |||
| - | + | == Advantages == | |
| + | |||
* No need to use restriction enzymes: this can sometimes be very tricky, if a cleavage site is present in one of your sequences. Also, it means that you can perform Gibson on any DNA sequence. | * No need to use restriction enzymes: this can sometimes be very tricky, if a cleavage site is present in one of your sequences. Also, it means that you can perform Gibson on any DNA sequence. | ||
* Simplified reaction: 45min at 50°C replaces digestions and religations. | * Simplified reaction: 45min at 50°C replaces digestions and religations. | ||
| Line 39: | Line 39: | ||
| - | + | == Disadvantages == | |
* Primers are more complicated to design and longer - they are more expensive and take longer to synthesize | * Primers are more complicated to design and longer - they are more expensive and take longer to synthesize | ||
* The reaction mastermix is expensive - due to large amounts of Taq ligase needed | * The reaction mastermix is expensive - due to large amounts of Taq ligase needed | ||
* A lot of negative resluts, du to the tendency of the backbones to close up themselves | * A lot of negative resluts, du to the tendency of the backbones to close up themselves | ||
Revision as of 07:23, 12 September 2011
Gibson assembly
Gibson assembly is a useful DNA ligation method that offers an alternative to the classical approach of using restriction enzymes. The procedure was was developed in 2009 by Gibson et al [1] and was introduced to the iGEM competition by the Cambridge 2010 team.
We successfully used Gibson assembly for our plasmid assemblies, allowing us to put different genes in our plasmid backbones. You can find our protocol in our Notebook, under "Protocols" section.
Contents |
Requirements
In order to perform Gibson assembly on two DNA sequences, these sequences need to overlap by at least 20bp at the edge. You can easily add these overlaps by extension PCR
Principle
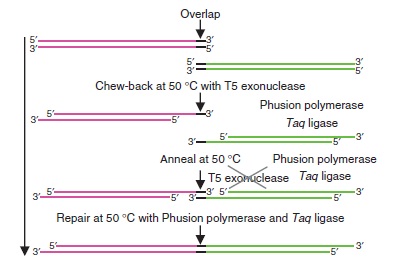
Gibson assembly relies on the activity of 3 different enzymes:
- T5 exonuclease
- Phusion polymerase
- Taq ligase
Combined together, these 3 enzymes allow overlapping DNA sequences (sequences A and B) to be ligated together in a single and isothermal reaction. First, the T5 exonuclease chews up bases at the 5' end on each strand, creating single-strands on the edges. Due to the sequence overlap, the single strand from A will anneal with the single strand from B. Then, the Phusion polymerase extend the DNA sequence from the 3' of the AB overlap. Finally, when there all the gaps have been filled by the polymerase, the Taq ligase ties up the A and B fragments.
Step-by-step
- Preparing the parts: For every pair of parts that you want to asemble, you need to add 20bp overhang with the other part. This implies running a PCR with primers that contain the overhang on their 5' end. After the PCR, you can purify your reaction products with a PCR purification kit and measure the DNA concentration.
- Preparing the assembly reaction: In order to have a successful reaction, you must mix together equimolar ratios of each of the parts. An easy way to calculate this is to divide the DNA concentration by the part length; you can also go on the Gibthon Construct Designer website and use the online tool. For inserting a gene in a plasmid backbone, we suggest to use a 2:1 ratio of the gene over the backbone.
- Running the reaction: Add Gibson mastermix and simply heat the tubes at 50°C for 45 minutes. You can use a normal PCR cycler for this step.
- Transforming cells: After the reaction, simply use a few microliters of it to transform cells.
- Negative control: For the negative control, make a Gibson reaction on just the plasmid backbone DNA and transform it. This will show you how much of the backbone closes itself up in your actual Gibson reaction.
Advantages
- No need to use restriction enzymes: this can sometimes be very tricky, if a cleavage site is present in one of your sequences. Also, it means that you can perform Gibson on any DNA sequence.
- Simplified reaction: 45min at 50°C replaces digestions and religations.
- No scar: this allows you to place a promoter and RBS in front of a gene for example
Disadvantages
- Primers are more complicated to design and longer - they are more expensive and take longer to synthesize
- The reaction mastermix is expensive - due to large amounts of Taq ligase needed
- A lot of negative resluts, du to the tendency of the backbones to close up themselves
 "
"