Team:EPF-Lausanne/Our Project/TetR mutants
From 2011.igem.org
(Difference between revisions)
(→Background information about TetR) |
|||
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:EPF-Lausanne/Templates/ | + | {{:Team:EPF-Lausanne/Templates/TetRtemplate|title=''In-Vitro'' Characterization}} |
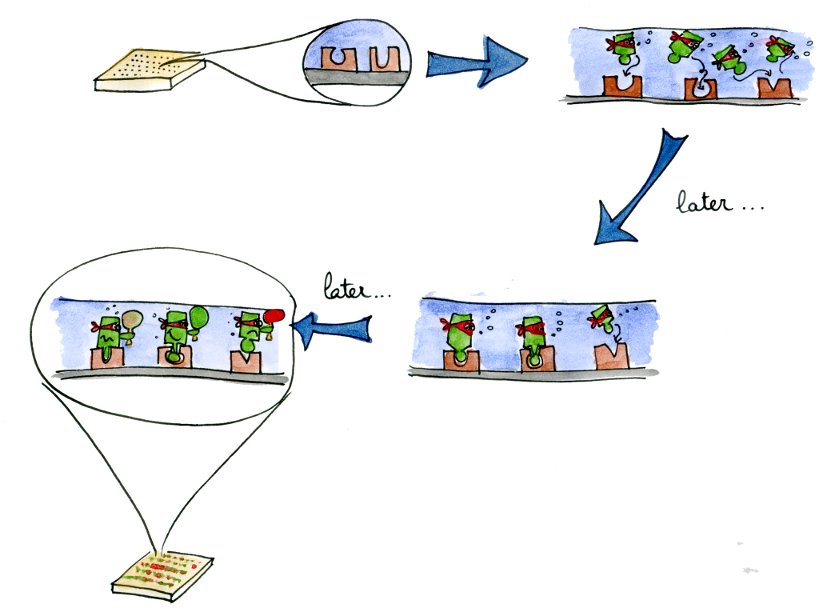
| - | + | When we generate new libraries of transcription factors using site-directed mutagenesis, we need to have a precise and high-throughput characterization method. These two requirements are met by the microfluidics MITOMI device that we used. | |
| - | + | ||
| - | + | Characterizing a lot of different mutants will allow us to get a better understanding of the mutations influencing the specificity and affinity of a transcription factor. With these parameters in hand, we will be able to fine-tune the characteristics of the newly produced transcription factors. | |
| - | + | The new transcription factors mutants characterized in vitro then need to be tested ''in vivo''. This last step is explained in the " ''in vivo'' characterization section". | |
| - | + | [[File:EPFL-Solange-MITOMI.jpg|700px]] | |
| - | + | ||
| - | [[File: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 02:59, 22 September 2011
In-Vitro Characterization
In vitro Main | Why TetR? | Mutant TetRs | MITOMI Data | In-vivo & In-vitro outlineWhen we generate new libraries of transcription factors using site-directed mutagenesis, we need to have a precise and high-throughput characterization method. These two requirements are met by the microfluidics MITOMI device that we used.
Characterizing a lot of different mutants will allow us to get a better understanding of the mutations influencing the specificity and affinity of a transcription factor. With these parameters in hand, we will be able to fine-tune the characteristics of the newly produced transcription factors.
The new transcription factors mutants characterized in vitro then need to be tested in vivo. This last step is explained in the " in vivo characterization section".
 "
"