Team:Colombia/Notebook/Plasmids
From 2011.igem.org
Template:Https://2011.igem.org/User:Tabima
Contents |
Plasmids
The Plasmids will be constructed as shown below

Plasmid 1
Plasmid 1 mission is to recognize the presence of chitin and to up regulate the promoter of the chitinolitic operon when chitin is abundant in the media. We´ve extracted and amplified chitoporin, chitin sensor and chitin binding protein (CBP) from Vibrio fischerii and aimed to create the bricks needed for the assembly of the plasmid that can recognize and break the chitin association.
Plasmid 2
Given that the chitin detection process is always active due to its basal expression, it is necessary to have a regulation so that the response signal to the plant could have two levels of expression. If the response signal to the plant is not regulated, it is possible to have fake or erroneous defense system activation signals when they are not needed, inducing resource losses that should be directed to plant normal growth and fruit production. It is therefore optimal to have the the signaling system activated only when there is a constant chitin presence.
The mechanism is planned to have two parts: a positive feedback that transforms the graduated input signal to an on/off signal with time delay, and an activation mechanism that requires both the delayed signal and the direct chitin signal, so that the system activates only when there is a constant time lapse with phytopatogen presence.
Positive Feedback
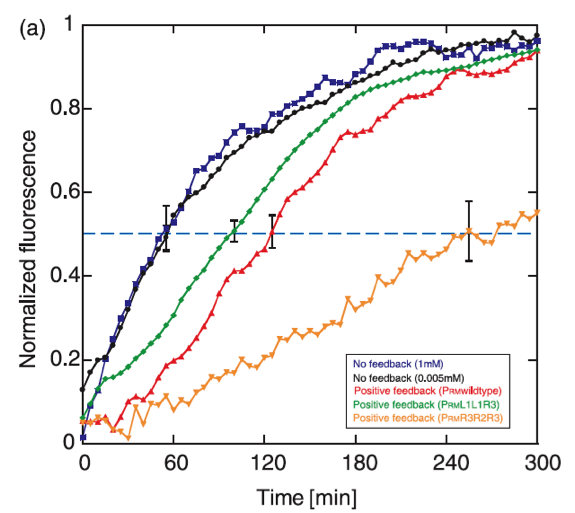
The positive feedback genetic system is well studied, and shows theoretically as well as experimentally a bistable response, moving between two discrete stable states without being able to stay in intermediate states. Another property that exhibits this type of feedback is the capability to delay the protein synthesis kinetics.
The operon chosen to control the response is based on the promoter PRM and the cI gene from the lambda bacteriophage. In this system, CI positively regulates its own synthesis, and the feedback activation guarantees the existance of two stable levels of expression. It has been found that the rise time of the positive feedback system expression is activated slower than a system without feedback, with delay times of approximately 70 minutes.
The dynamics of the graduated response of the positive feedback system shows also hysteretic propierties, which is useful in the signaling system to the plante, keeping the response active for an additional time after the phytopatogen is attacked with chitinase, so that the immunity is not lost when there is a chance of infection.
Mechanism of plant signaling activation
The defense system activation singaling gene is regulated by a two promotor system: one that is activated by the positive feedback system output (graduated and delayed signal) and another one that is activated directly by the presence of chitin (phytopatogen signal). The signal to the plant would be in its maximal expression only when the two input signals are active, which means in practical terms that only when there is a phytopatogen presence superior to 70 minutes the signal is sent, avoiding unnecesary resource expenses by the plant, for example, when an insect passes by the leaves. The hysteretic dynamics of the system has an additional advantage: the inmune response and the chitinase production (which attacks directly the phytopatogen) would be active for an additional time after the critical phytopatogen concentration is reached
Complete genetic circuit desing
In the figure the two planned mechanisms are shown. The first one is the positive feedback system, and its graduated, delayed output is the input for the second one, which is in charge of expressing the chitinase to attack the phytopatogen, and sending the input signal for the plant inmune system activation.
Plasmid 3
In a nutshell, P3’s mission is that of interpreting P2’s autoinduction signals (LuxI) in a way so that it responds with an appropriate production of the plant’s Systemic Aquired Resistance (SAR) stimulating factor (Salicylic Acid, SA). We have also included a negative feedback loop in order to control possible gene overexpressions by introducing a LuxI activated lactonase.
Test Plasmid
Building a whole working system is the challenge our group faces everyday. But, as important as the assemblage itself, testing our assumptions about how the designed parts work individually, helps us to improve the molecular design. Vibrio's chitin detection system relies on the activation of the promoter of the chi genes by a [http://www.pnas.org/content/101/2/627.full two-component signaling device]. Since the sequence of the promoter remains uncharacterized, we extracted a small region of the sequence upstream the chitoporin gene (promoter chiP) and used it as the promoter activated when chitin is present in the environment. To test whether this sequence acts as a promoter in the presence of chitin, we transformed an E. coli K12 with a vector containing GFP downstream this sequence, using GFP without promoter as control. This experiment is currently being performed.
The core and main idea of the whole Cropspirin system is to warn the coffee plant when a potential fungal pathogen lies on its surface. A way of conveying the message of "alarm", is to use plant hormones like [http://books.google.com/books?id=AxUhFMcwSrMC&printsec=frontcover&hl=es#v=onepage&q&f=false Salicylic acid]; this hormone can elicit an immune response known as Systemic Acquired Resistance (SAR) which means that the plant is in a defense state, therefore will be resistant to the majority of infections. To test our system, we first need to probe if the [http://partsregistry.org/wiki/index.php/Part:BBa_J45320 Salycilate generator (MIT iGEM 2006)] is able to induce plant defense responses in vivo. In order to do that we used two different approaches:
1. We challenged Arabidopsis thaliana Col 0, a well studied model plant, with a IPTG induced K12 strain carring either [http://partsregistry.org/wiki/index.php/Part:BBa_J45320 Salycilate generator (MIT iGEM 2006)] or the empty backbone at different cell densities (1x10 3, 1x10 5, 1x10 6). We harvest the leaves 48h psi and extracted RNA to look for differences in the expression of [http://www.pnas.org/content/104/25/10732 PR-1], a protein expressed when the plant defense is induced. We are currently performing the RT-PCR and expecting for good results.
2. We also used our plant pathogen model of study Coffea arabica-Hemileia vastatrix to test if the immunization mediated by [http://partsregistry.org/wiki/index.php/Part:BBa_J45320 Salycilate generator (MIT iGEM 2006)] in coffee plants can avoid the development of the rust infection. For this purpose we inoculate by spraying or infiltrating small susceptible plants of coffee with the IPTG induced K12 strain carrying either [http://partsregistry.org/wiki/index.php/Part:BBa_J45320 Salycilate generator (MIT iGEM 2006)] or the empty backbone at different cell densities (1x10 3, 1x10 5, 1x10 6. 1 week psi we challenge the plants with a rust spores suspension. We are currently expecting the non development of symptoms.
 "
"