Team:Calgary/Project/Project/Chassis/Microalgae
From 2011.igem.org
Emily Hicks (Talk | contribs) |
Rpgguardian (Talk | contribs) |
||
| Line 7: | Line 7: | ||
<h2>Microalgae Tools</h2> | <h2>Microalgae Tools</h2> | ||
| - | <p>In | + | <p>In order to be able to work efficiently with microalgae, protocols for various procedures were needed. We wanted to eventually use our microalgae strain to construct a promoter library out of nuclear and chloroplast DNA. As such, transformation as well as nuclear DNA and chloroplast extraction protocols were necessary. Unfortunately, our strain of microalgae, <i>Dunaliella tertiolecta</i>, does not have well established transformation protocols defined in the literature, and as such time was spent optimizing literature protocols. All microalgae protocols we used can be found in our <a href="https://2011.igem.org/Team:Calgary/Notebook/Protocols">protocol section. </a> </p> |
<h2>Transformation</h2> | <h2>Transformation</h2> | ||
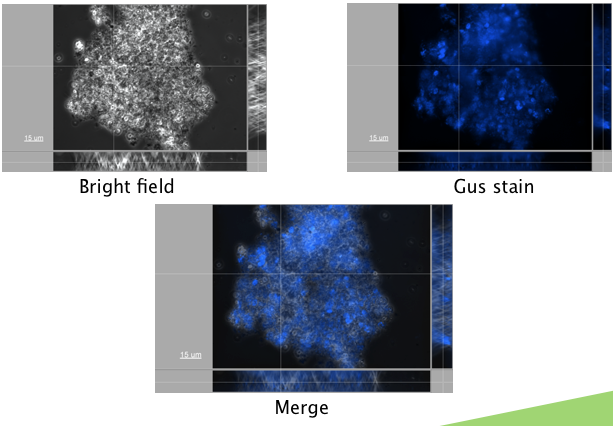
| - | <p>An appropriate reporter also needed to be found. A reporter construct containing GFP downstream of a Nopaline Synthase (NOS) promoter was first transformed. Autofluorescence levels were found to be too high | + | <p>An appropriate reporter also needed to be found. A reporter construct containing GFP downstream of a Nopaline Synthase (NOS) promoter was first transformed. Autofluorescence levels were found to be too high to detect fluorescence above the background, making this a poor choice for a reporter. A GUS reporter commonly used in plants, and available in the registry (<a href="http://partsregistry.org/Part:BBa_K330002">BBa_K330002</a>) was attempted. GUS (beta-glucuronidase) is an enzyme from <i>E. coli</i>, that is frequently used a s a plant reporter. A construct containing the GUS reporter gene, downstream of the NOS reporter was successfully transformed (figure x.)</p> |
| - | </html>[[Image:Calgary2011_Microalgae_Transformation_Picture.png|thumb|600px|center| | + | </html>[[Image:Calgary2011_Microalgae_Transformation_Picture.png|thumb|600px|center|Transformation of <i>Dunaliella tertiolecta</i> with a GUS containing plasmid with a NOS promoter. Expression was measured at 500 nm and bright field view. Resulting images were merged showing successful transformation of the microalgae plasmid.]]<html> |
| - | |||
<h2> New Microalgae Parts</h2> | <h2> New Microalgae Parts</h2> | ||
| - | Looking through the registry, | + | Looking through the registry, our team recognized that for future teams to use microalgae effectively, there were several parts that would be required. The registry contains a few appropriate selectable markers (kanamycin and gentamycin), a strong constitutive 35S CaMV promoter (BBa_K509000), as well as a visual GUS reporter (BBa_K330002). What was needed was an inducible promoter and a more quantitative reporter gene. To this end, we biobricked the Hsp70A/RbcS2 promoter from <i>Chlamtdomonas reinhardtii</i> (BBa_K640001), which is inducible by light/ heat shock. We also biobricked a gene for a codon optimized algal luciferase. Both genes were obtained from the Clamydomonas Database, an online registry for chlamydomonas tools. |
| - | |||
| + | <h2> Characterization of the Hsp70A/RbcS2 promoter (BBa_K640001)</h2> | ||
| - | + | Characterization data was collected for a microalgae construct containing both the Hsp70A promoter attached to the algal luciferase. Constructs were transformed with our glass bead method and allowed to incubate at various temperatures (<b>put in the termpatures here</b>) | |
| + | </html>[[File:UPutinyourfilenamehere]][[File:UCalgary2011 Algae Luciferase.png]]<html><br><br> | ||
Chlamydomonas reibhrdtii cells were transfored with a plasmid DNA of a construct containing the Hsp70A/RbcS2 promoter upstream of luciferase. | Chlamydomonas reibhrdtii cells were transfored with a plasmid DNA of a construct containing the Hsp70A/RbcS2 promoter upstream of luciferase. | ||
</html> | </html> | ||
}} | }} | ||
Revision as of 23:58, 28 September 2011









A Chassis Using Native Tailings Pond Species

Microalgae Tools
In order to be able to work efficiently with microalgae, protocols for various procedures were needed. We wanted to eventually use our microalgae strain to construct a promoter library out of nuclear and chloroplast DNA. As such, transformation as well as nuclear DNA and chloroplast extraction protocols were necessary. Unfortunately, our strain of microalgae, Dunaliella tertiolecta, does not have well established transformation protocols defined in the literature, and as such time was spent optimizing literature protocols. All microalgae protocols we used can be found in our protocol section.
Transformation
An appropriate reporter also needed to be found. A reporter construct containing GFP downstream of a Nopaline Synthase (NOS) promoter was first transformed. Autofluorescence levels were found to be too high to detect fluorescence above the background, making this a poor choice for a reporter. A GUS reporter commonly used in plants, and available in the registry (BBa_K330002) was attempted. GUS (beta-glucuronidase) is an enzyme from E. coli, that is frequently used a s a plant reporter. A construct containing the GUS reporter gene, downstream of the NOS reporter was successfully transformed (figure x.)
New Microalgae Parts
Looking through the registry, our team recognized that for future teams to use microalgae effectively, there were several parts that would be required. The registry contains a few appropriate selectable markers (kanamycin and gentamycin), a strong constitutive 35S CaMV promoter (BBa_K509000), as well as a visual GUS reporter (BBa_K330002). What was needed was an inducible promoter and a more quantitative reporter gene. To this end, we biobricked the Hsp70A/RbcS2 promoter from Chlamtdomonas reinhardtii (BBa_K640001), which is inducible by light/ heat shock. We also biobricked a gene for a codon optimized algal luciferase. Both genes were obtained from the Clamydomonas Database, an online registry for chlamydomonas tools.Characterization of the Hsp70A/RbcS2 promoter (BBa_K640001)
Characterization data was collected for a microalgae construct containing both the Hsp70A promoter attached to the algal luciferase. Constructs were transformed with our glass bead method and allowed to incubate at various temperatures (put in the termpatures here) File:UPutinyourfilenamehere
Chlamydomonas reibhrdtii cells were transfored with a plasmid DNA of a construct containing the Hsp70A/RbcS2 promoter upstream of luciferase.

 "
"