Team:Calgary/Project/Preliminary Data
From 2011.igem.org
Emily Hicks (Talk | contribs) |
|||
| Line 12: | Line 12: | ||
</html>[[File:UCalgary2011 IMG 0528.JPG|300px]][[File:UCALGARY2011 ColorChange!.jpg|240px]]<html> | </html>[[File:UCalgary2011 IMG 0528.JPG|300px]][[File:UCALGARY2011 ColorChange!.jpg|240px]]<html> | ||
<br><br> | <br><br> | ||
| - | An | + | An electrode in a CPR solution is shown on left, while a CPRG solution is shown on the right. |
<br> | <br> | ||
<br> | <br> | ||
Revision as of 02:47, 29 September 2011









Preliminary Testing

Background and Rationale
For the purposes of testing the lacZ reporter gene (BBa_I732005) as an electrochemical detector it was important to first assess how well the reporter performed at cleaving the chlorophenol red β-D-galactopyranoside (CPRG) into Chlorophenol red (CPR). LacZ downstream of an IPTG inducible promoter(lacI) was used, already present in the registry as BBa_I732901. The purpose of the experiment was to ensure that the E. coli could be induced to produce β-galactosidase, which could then cleave chlorophenol red β-D-galactopyranoside (CPRG) to produce Chlorophenol red (CPR). Since CPR has a very dark characteristic red color at pH 7 the reaction was to be kept at that constant pH using a phosphate buffer. This experiment was the first step in characterizing lacZ as an electrochemical reporter (i.e. oxidizing the CPR and measuring either the current or voltage). We hoped it would not only show that our cells could produce enough beta-galactosidase to produce a detectable amount of CPR, but that it could also help in determining what sort of time frame would be needed for the E. coli to produce a significant amount of CPR by cleavage of CPRG providing a guideline for future electrochemical testing.


An electrode in a CPR solution is shown on left, while a CPRG solution is shown on the right.
Methods
E. coli carrying the plasmid were cultured until log phase then exposed to PBS buffer containing 3mM CPRG and 1mM IPTG for fixed lengths of time before measuring the absorbance of the solution. A detailed procedure are recorded in protocols section.
Results
Discussion of Results
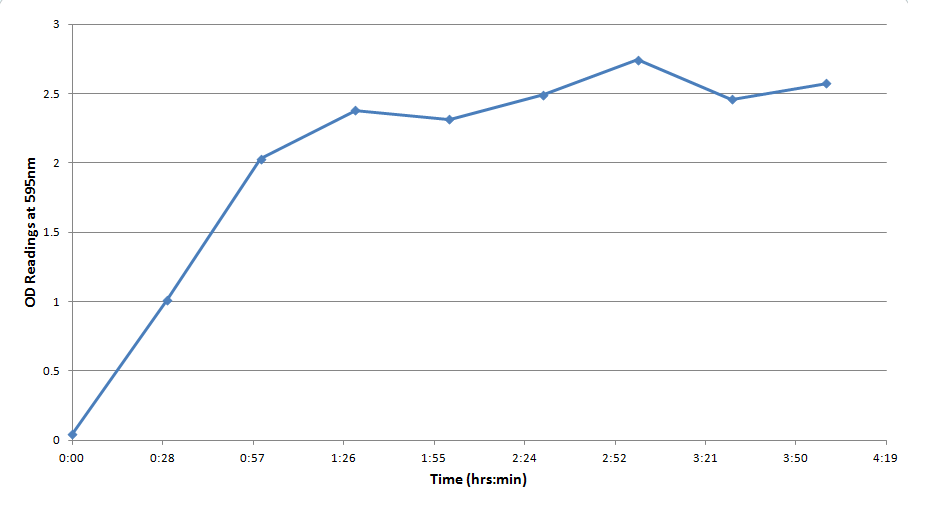
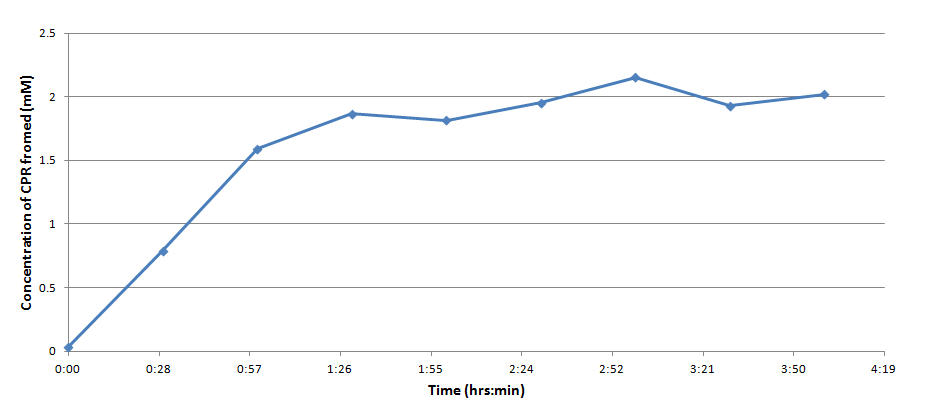
Figure 3 indicates that the concentration of CPR clearly increases over time in this mixture. Illustrating that the IPTG is successfully inducing the lacZ gene to produce β-galactosidase. This data will be useful to the electrochemical detector side of the project because they will now have data from which they can choose a time when they should begin oxidizing the CPR formed by the E. coli (by cleavage of CPRG).
One drawback experienced in this experiment was that the spectrophotometer which was used to take optical density readings could not give data past an OD of ~3 for CPR absorption, meaning that even samples with an extremely high concentration of CPR would at best have an absorbance reading similar to a ~3 mM sample. Thus, this method was not particularly accurate for highly concentrated solutions of CPR. However the results did reveal that under the specified conditions about 1mM of CPR could be produced in around 30 minutes revealing that the system responded quite rapidly when induced with IPTG.

 "
"