Team:Calgary/Project/DataPage
From 2011.igem.org
(Difference between revisions)
Rpgguardian (Talk | contribs) |
Rpgguardian (Talk | contribs) |
||
| Line 20: | Line 20: | ||
<h2>Additional Work and Characterization</h2> | <h2>Additional Work and Characterization</h2> | ||
| - | <ul><li><b>Bba_K300022</b> - Optimized this part for transformation into microalgae (<a href="https://2011.igem.org/Team:Calgary/Project/Project/Chassis/Microalgae">See microalgae | + | <ul><li><b>Bba_K300022</b> - Optimized this part for transformation into microalgae (<a href="https://2011.igem.org/Team:Calgary/Project/Project/Chassis/Microalgae">See microalgae section</a>) |
| - | <li>Characterized the | + | <li><b>Bba_I732901</b> - Characterized the breakdown of CPRG to CPR using spectrometry over time (<a href="https://2011.igem.org/Team:Calgary/Project/Preliminary_Data">See reporter characterization section</a>) |
</ul> | </ul> | ||
Revision as of 03:26, 29 September 2011









Building a Naphthenic Acid Biosensor

The Design
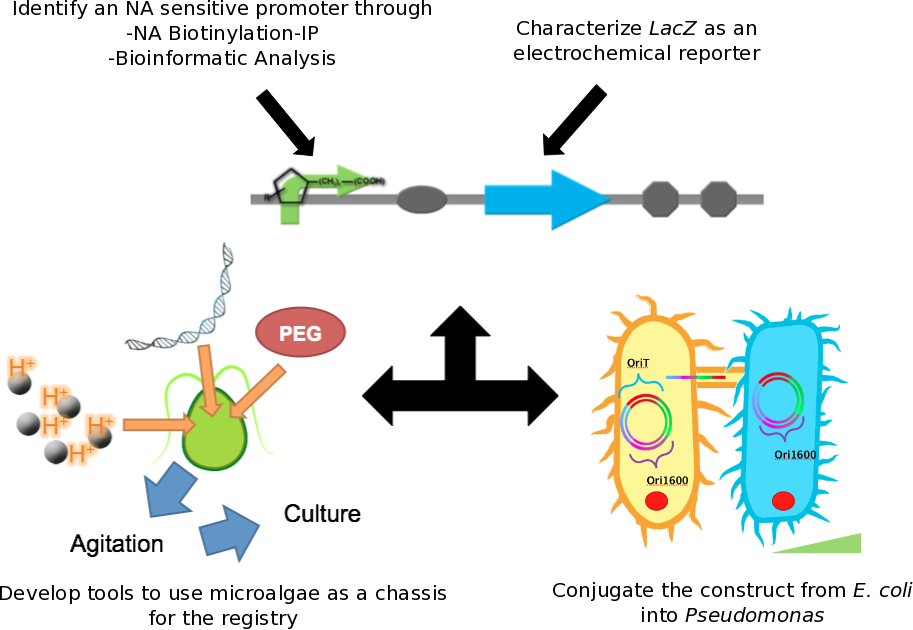
Figure 1 In order to develop a naphthenic acid biosensor we were required to develop a sensory element which would respond specifically to this class of compounds. Additionally, we developed a novel electrochemical reporter system using the lacZ gene product Beta-galactosidase to detect and quantify the response of our promoter into a measurable system. Since E. coli is not naturally found in tailings ponds, we designed a novel conjugation system to transfer our reporter system into Pseudomonas as well as protocols for use of our system in microalgae.
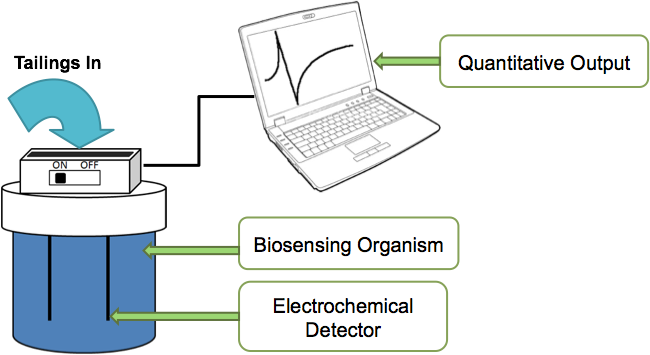
Figure 2 Our vision was to use our construct in a device which could be used to quantitate the amount of naphthenic acids in a tailings pond solution. A sample would be loaded into the device, the bacteria would respond to the naphthenic acids in solution and produce an electrochemical output. This could be quantitatively measured and the output graphed onto a computer.
Data From New Parts Submitted To The Registry
- Bba_K640002, Bba_K640003, Bba_K640004, Bba_K640005 - These parts contain oriT, ori1600, oriT-ori1600, and oriT(Heidelberg Part)-ori1600 respectively. We have characterized the expression of these parts on our Pseudomonas chassis section of the wiki.
- Bba_K640000, and Bba_K640001 - Codon optimized Luciferase and Hsp70A/RbcS2 Microalgae parts. More information can be found in our microalgae chassis section.
Data From Characterized Parts Already In the Registry
- Bba_I732005 - Characterized the LacZ gene as an electrochemical detector using chlorophenol red. Please see the reporter section of our wiki and associated characterization page for more information.
Additional Work and Characterization
- Bba_K300022 - Optimized this part for transformation into microalgae (See microalgae section)
- Bba_I732901 - Characterized the breakdown of CPRG to CPR using spectrometry over time (See reporter characterization section)

 "
"






