Team:Calgary/Project/Chassis
From 2011.igem.org
(Difference between revisions)
Emily Hicks (Talk | contribs) |
|||
| (3 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
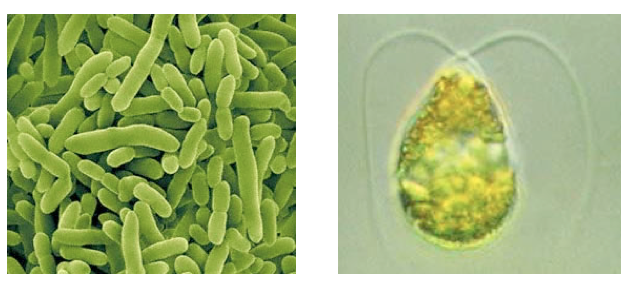
</html>[[Image:Univeristy_of_Calgary2011_Chassis_Visuals.png|frame|center|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> | </html>[[Image:Univeristy_of_Calgary2011_Chassis_Visuals.png|frame|center|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> | ||
<h2>Chassis Selection</h2> | <h2>Chassis Selection</h2> | ||
| - | We identified early on in our project planning stages that we would like to use <i>Pseudomonas</i> or microalgae as the chassis to house our system. | + | We identified early on in our project planning stages that we would like to use <i>Pseudomonas</i> or microalgae as the chassis to house our system. The motivation for this is that not only are these organisms capable of naturally surviving a tailings pond environment, but they may contain necessary regulatory elements required for a response to NAs. Because of this, taking a response element out of an organism and putting our system into another chassis, such as <i>E. coli</i>, may not behave as expected. Additionally, we characterized a change in growth rate of <i>E. coli</i> with and without tailing ponds showing that, cells don't grow as well in this media (This work was done in collaboration with the Lethbridge team - See our <a href="https://2011.igem.org/Team:Calgary/Outreach/Collaboration">collaboration section</a> for more information on this research). Based on this chassis choice however, we realized that given their scarcity in iGEM, tools were needed to establish work in these organisms for future iGEM teams. |
<br><br> | <br><br> | ||
| Line 13: | Line 13: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | <a href="https://2011.igem.org/Team:Calgary/Project/Chassis/Pseudomonas">Click here</a> to read more about our Pseudomonas Tools. | + | <a href="https://2011.igem.org/Team:Calgary/Project/Chassis/Pseudomonas">Click here</a> to read more about our <i>Pseudomonas</i> Tools. |
<style> | <style> | ||
Latest revision as of 04:38, 29 September 2011









A Chassis Using Native Tailings Pond Species

Chassis Selection
We identified early on in our project planning stages that we would like to use Pseudomonas or microalgae as the chassis to house our system. The motivation for this is that not only are these organisms capable of naturally surviving a tailings pond environment, but they may contain necessary regulatory elements required for a response to NAs. Because of this, taking a response element out of an organism and putting our system into another chassis, such as E. coli, may not behave as expected. Additionally, we characterized a change in growth rate of E. coli with and without tailing ponds showing that, cells don't grow as well in this media (This work was done in collaboration with the Lethbridge team - See our collaboration section for more information on this research). Based on this chassis choice however, we realized that given their scarcity in iGEM, tools were needed to establish work in these organisms for future iGEM teams.Click here to read more about our Microalgae Tools.
Click here to read more about our Pseudomonas Tools.

 "
"