Team:BU Wellesley Software/Puppetshow
From 2011.igem.org
PuppetShow
Tool Overview
Laboratory protocols often require careful and precise execution of numerous procedures to achieve desired results. However, slight inaccuracies and miscalculations attributable to human-error continually arise and often lead to invalid conclusions. For this reason, we have developed an accurate workflow that executes biological protocols in a more systematic manner. The solution we propose is the Puppeteer Biological Protocol Automation Suite. Our suite centers on the execution of Puppeteer, a high-level protocol specification language used to programmatically describe biological protocols; it is executed on a liquid-handling robot with minimal user intervention. PuppetShow allows users to create protocols locally, but will eventually incorporate a “Protocol Repository” that will retain version-controlled instances of uploaded protocols. This feature will promote all forms of collaboration within the synthetic biology community, with a predominant focus on inter-laboratory protocol sharing.
Puppeteer Assembly Flow
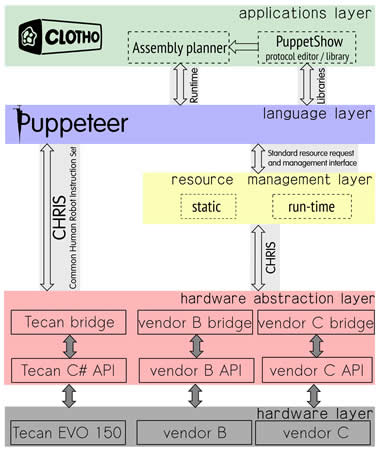
Our assembly flow incorporates a five-layer stack as illustrated in the adjacent figure. The Assembly Planner produces an assembly plan for synthetic biological devices, with each assembly step annotated with the name of a biological protocol. Each protocol may be fully specified using PuppetShow.
The protocols are written in the high-level language called Puppeteer. The Language layer includes a Puppeteer interpreter and linker. A protocol specified in Puppeteer may contain Puppeteer instructions as well as references to previously created Puppeteer programs available in the protocol repository. The Language layer then translates Puppeteer protocols into low-level commands expressed in a Common Human Robot Instruction Set (CHRIS). CHRIS can be produced by any high-level language, making it ideal for robot vendors to utilize. The Hardware Layer concerning the external control and robot I/O interfacing is wrapped under a Hardware Abstraction Layer (HAL). Vendor-provided software for programming the robot may be proprietary and is used to control the robot. An interface to it is provided by a software bridge, which maps protocols expressed in CHRIS to sequences of native robot instructions.
The Resource Management layer maintains resource state information and allows for high-level interface standardizations for initializing, requesting, naming, aggregating, and accessing resources to the Language layer, analogous to a ''system call'' suite. This interface abstracts resource management away from the protocol specification language.
Demo
Software Demo Video
Live Robot Demo Video
Results
An initial version of the Puppeteer stack is fully integrated with the Clotho software platform and implemented within the wet lab. Basic BioBricks assembly protocols have been implemented and validated via the physical assembly of basic devices. Two protocols central to BioBricks assembly—Restriction Digestion and Ligation—have been created using the Puppeteer language. The Puppeteer implementation was validated by executing multiple trials for both protocols and verifying the results via running gels.
Collaboration with other iGEM teams:
In order to ensure reproducibility and promote collaboration, we visited the Weiss lab at MIT to implement the Puppeteer stack with their robot configuration. During the visit we encountered a few MIT iGEM team members and coordinated with them to test the Puppeteer flow with the MIT Tecan robot. After we got settled, we noticed several differences between MIT's and our own Tecan robot deck setups; this required modifying settings within the Puppeteer flow for proper protocol execution. As such, we separated modifiable settings into an easy-to-edit settings file to ensure that Puppeteer maintains configurable settings for varying deck layouts. The Puppeteer flow was then tested on MIT’s robot, and everything proceeded as expected. The entire collaborative experience with MIT’s iGEM team remains fruitful since it enforced a more robust and functional Puppeteer flow across multiple robot configurations.
Restriction Digest:
 For restriction digest we digested the plasmid containing the BioBrick BBa_J52028 in order to isolate the GFP. After cutting the plasmid with EcoRI and SpeI we ran it on a one percent gel along with a ladder in order to determine the amount of base pairs for each DNA part. As GFP is about 1221 bp the location in which it is located on the gel is correct. Also the backbone was about 3189 bp which is also located in the correct location on the gel.
For restriction digest we digested the plasmid containing the BioBrick BBa_J52028 in order to isolate the GFP. After cutting the plasmid with EcoRI and SpeI we ran it on a one percent gel along with a ladder in order to determine the amount of base pairs for each DNA part. As GFP is about 1221 bp the location in which it is located on the gel is correct. Also the backbone was about 3189 bp which is also located in the correct location on the gel.
Ligation:
Our first ligation involved BBa_J23100 (RFPc) and BBa_I14033(Pcat). The BBa_J23100 was cut with SpeI and PstI in order to remove the RBS-RFP-Terminator composite part. Three trials were run on the robot with the same parts and the same amounts. A fourth trial was run manually alongside the robot. When ligations were done the total amount for each robot trial was about 5ul off from the desired amount. It took one day for the colonies to grow on the plates but it took about two days for them to finally turn a pinkish color. The ligation was restriction mapped in order to verify that the robot worked properly. One of the robot trials had good results but we needed more verification in order to state that it was successful. Also there were very few colonies so the process had to be fixed.
We tried ligating again but this time we used BBa_J23101 with Pcat, as we did not have enough digested BBa_J23100. The colonies were very faint and did not seem to produce good outcomes. We decided to test another promoter in order to continue forward. BBa_R0040 (Ptet) worked better so we did two trials on the robot with BBa_J23100 and BBa_R0040 and then one manually. This time around, the colonies looked to be the right color but there were still not enough colonies to call the outcome a success. We decided to take a different approach for fixing the issue by testing new parts. We did a ligation using large parts that have had good ligation outcomes manually. The parts were ligated so that the order in the plasmid is Ptet-RBS-LuxR-YFPc-pLuxR. After being transformed on Ampicillin plates there were a lot of colonies on both the robot trials and the manual trials. This can be seen verified in following figure.

 We also did a double volume ligation at the same time as the Ptet-RBS-LuxR-YFPc_pLuxR ligation to see if we could fix the volume issue. The final amounts for each ligation this time was only off by 4ul. BBa_E0240 (GFPc) and promoter BBa_R2000 were used. Unfortunately no colonies grew on the plates. The ligations for Ptet-RBS-LuxR-YFPc-pLuxR was tested by restriction mapping the part. Colony A did not ligate properly but colony B and C did work. The insert is about the same size as the backbone so the fact that there are two bands right under each other is correct. As there was no control for the first double volume ligation, we repeated it but we used RFPc and Ptet once again and did a single volume ligation at the same time. This time the plates clearly showed that the double ligation had the same, if not more, colonies on the plates than the manual control. The regular ligations with the RFPc and Ptet unfortunately still had discrepancies between the robot trials and the manual trial. The restriction mapping for the single and double volume ligations proved that both ligations worked properly.
We also did a double volume ligation at the same time as the Ptet-RBS-LuxR-YFPc_pLuxR ligation to see if we could fix the volume issue. The final amounts for each ligation this time was only off by 4ul. BBa_E0240 (GFPc) and promoter BBa_R2000 were used. Unfortunately no colonies grew on the plates. The ligations for Ptet-RBS-LuxR-YFPc-pLuxR was tested by restriction mapping the part. Colony A did not ligate properly but colony B and C did work. The insert is about the same size as the backbone so the fact that there are two bands right under each other is correct. As there was no control for the first double volume ligation, we repeated it but we used RFPc and Ptet once again and did a single volume ligation at the same time. This time the plates clearly showed that the double ligation had the same, if not more, colonies on the plates than the manual control. The regular ligations with the RFPc and Ptet unfortunately still had discrepancies between the robot trials and the manual trial. The restriction mapping for the single and double volume ligations proved that both ligations worked properly.
Safety practices
As wet-lab protocols were repeated on the robot the same safety procedures were performed. The trials that were run on the robot were done inside a closed laboratory meant for biological processes. Also while the robot was performing its task, individuals were not allowed to interfere directly with the robot.
None of the materials used in our iGEM project pose a significant risk to the health and safety of our team members and laboratory. We work in a standard BSL1 certified lab space and mainly work with ''E. coli'' bacteria. Furthermore, there is no risk to the public health or environment if the materials of our project were released. This is because the genes that we are using and studying code for transcriptional proteins and are not pathogenic or toxic. There is no threat of security through misuse of our materials because of their nonlethal nature. In terms of lab safety and institutional standards, there is an Institutional Biosafety Committee (IBC) at Boston University which evaluates all research projects, primarily those involved with recombinant DNA, and insures proper biosafety guidelines. We are in contact with the IBC and are in the process of approving our project.
There is also a Laboratory Safety Committee through which all wet lab members have taken mandatory laboratory safety training. This training provides a basic overview to chemical and biosafety, fire and life safety, emergency management, and waste management. In the United States, the major centers for biosafety procedures and guidelines are the National Institutes of Health (NIH) Guidelines and the Center for Disease Control and Prevention (CDC).
Future Work
-
Sample History Viewer
- Samples currently represent any liquid-like substance used within the laboratory environment. With the current working database implementation, output sample (e.g., a restriction digested part) information is stored with associated history. This sample history comprises the individual samples that were utilized to create the output sample (i.e., the buffers and enzymes, etc.) as well as the associated protocols that were executed. This entire flow has been implemented but is missing a viewer component that will allow users to visually explore the different histories of samples within the database.
-
Protocol Graph Editor
- The concept of a protocol graphs focuses on an ideal and simplistic hierarchical representation of biological protocols. The graph itself is a directed bipartite graph composed of protocol nodes, sample nodes, and connection arcs. The bipartite label implies that protocol nodes can only connect to sample nodes and vice versa (sample nodes are input(s)/output(s) to/from protocol nodes). With such a representation, complex biological protocols can be translated into a (potentially large) but scalable and useful graphical representation.
- Avi Robinson-Mosher, a postdoctoral researcher in the Silver lab at the Wyss Institute for Biologically Inspired Engineering at Harvard University, has shown interest in a protocol graph editor. We are collaborating with the Silver lab over data structures and database organization regarding the implementation and storage of protocol graphs.
-
Integration with eLabNotebook
- The eLabNotebook application uses a protocol representation to display procedural steps for wet lab convenience. Ideally, a layer of communication will coexist between PuppetShow’s database features and the eLabNotebook’s central server configuration. Generated protocol graphs and associated reports, created by users in Trumpet or any software tools created by future iGEM teams, will be stored on an identifiable server. The eLabNotebook will then pull and parse the information from the server to provide accurate, up-to-date information for the laboratory environment, thanks to PuppetShow's ability to transfer data between applications.
-
Integration with Trumpet
- Trumpet already generates reconfigurable constructs in the form of Clotho “composite parts”. This data model is already adaptable to the Puppeteer assembly flow, so it will be very feasible to execute protocols from Trumpet’s output. This feature can be implemented as soon as a standard for implementation is finalized.
-
Protocol optimizer project
- A potential project proposal involves the algorithmic assembly of protocols. This procedure would be completed on an abstract level, where a sophisticated evaluation of available resources is used to generate optimized protocols. The available resources would comprise samples within the database, both previously-created and newly-entered, that are paired up with protocols to provide efficient output creation. The ingenuity behind this application involves the usage of historical context, which is retained in the form of sample history within the database.
 "
"
