Team:Potsdam Bioware/Labjournal/June
From 2011.igem.org
10th Labday 2011-06-01
Primer design for Phage Display
Investigators: Jessica, Sabine, Sandrina, Nadja
Aim: Design primers for PCR of mdnA (from pARW089) to clone into pAK100 with SfiI restriction sites
Time: 2011-06-01,15:30-21:00
Materials:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
Primer design
- melting temperature based on 4+2 method (for mdnA-overlapping part)
- adding sequences for restriction sites (considering reading frame)
- Check self-complementarity and melting temperature using Oligo Calc
Results:
Primer sequences
- pf_sfi_mdna_1 TCTAGATGGCGGCCCAGCCGGCCATGGCGATGGCATATCCCAAC
- pr_sfi_mdna_1 CCCTTCTGACTGGGAAGATTATGGCCTCGGGGGCCACTAATACTAGTAGCGGCCGCTGCAGGCT
Further tasks:
- do PCR of mdnA
11th Labday 2011-06-07
Design of PCR conditions for amplifying the complete mdn cluster
Investigators: Jessica, Nicole, Nadja
Aim: PCR amplification of the mdn cluster
Time: 2011-06-07,18:00-19:30
Materials:
- Sequence of vector pARW071, concentration 857.3 ng/ µl
- Manual: Fermentas Long PCR Enzyme Mix
- Forward primer: sf_bb_4_1
- Reverse primer: r_bb_1
- Geneious Pro 5.1.7
Method:
- Design of a temperature profile based on the manual
- Definition of a mastermix based on the manual
Results:
1. Temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 180 | Initial denaturation |
| 94 | 15 | Denaturation |
| 46 | 30 | Annealing |
| 68 | 300 | Elongation |
| --> 10 cycles | ||
| 94 | 15 | Denaturation |
| 46 | 30 | Annealing |
| 68 | 300 + 2 per each cycle | Elongation |
| --> 20 cycles | ||
| 68 | 600 | Final extension |
2. Mastermix
| Component | Volume in µl | Final concentration |
|---|---|---|
| Buffer Fermentas Long Enzyme Mix (+MgCl2) | 5 | |
| dNTPs | 1 | 0.2 mM |
| Primer (each) | 3 | 0.6 µM |
| DNA (diluted 1:100) | 4.08 µl | 0.7 ng |
| Polymerase | 0.3 µl | 1.5 U |
| H2O | 33.62 µl | (up to 50 µl) |
Further tasks:
- PCR of the complete mdn cluster
- verification using agarose gel electrophoresis
Design of PCR conditions for amplifying mdnA
Investigators: Jessica, Nicole, Nadja
Aim: PCR amplification of mdnA
Time: 2011-06-07,18:00-19:30
Materials:
- Sequence of vector pARW089, concentration 626.7 ng/ µl
- Manual: NEB OneTaq DNA Polymerase
- Forward primer: sf_mdna_1
- Reverse primer: r_mdna_1
- Geneious Pro 5.1.7
Method:
- Design of a temperature profile based on the manual
- Definition of a mastermix based on the manual
Results:
1. Temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 30 | Initial denaturation |
| 94 | 15 | Denaturation |
| 45 | 35 | Annealing |
| 68 | 20 | Elongation |
| --> 30 cycles | ||
| 68 | 300 | Final extension |
| 6 | Hold |
2. Mastermix
| Component | Volume in µl | Final concentration |
|---|---|---|
| Buffer NEB OneTaq | 10 | |
| dNTPs | 1 | 0.2 mM |
| Primer (each, diluted 1:10) | 1 | 0.2 µM |
| DNA (diluted 1:100) | 5.58 | 0.7 ng |
| Polymerase | 0.25 | |
| H2O | 31.17 | (up to 50 µl) |
Further tasks:
- PCR of mdnA
- verification using agarose gel electrophoresis
12th Labday 2011-06-08
PCR of the complete mdn cluster
Investigators: Jessica, Nicole
Aim: Testing the possibility of a PCR of the whole mdn cluster and the chosen conditions
Time: 2011-06-08, 9:30-14:00
Materials:
- DNA polymerases: Long PCR Enzyme Mix (Fermentas)
- Template DNA: pARW071 (vector)
- Forward primer: sf_bb_4_1
- Reverse Primer: r_bb_1
--> mastermix composition, see 11th labday 'Design of PCR conditions for amplifying the complete mdn cluster'
- Eppendorf Mastercycler gradient
- used temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 120 | Initial denaturation |
| 94 | 20 | Denaturation |
| 56 | 20 | Annealing |
| 68 | 300 | Elongation |
| --> 10 cycles | ||
| 94 | 20 | Denaturation |
| 56 | 20 | Annealing |
| 68 | 300 + 2 per each cycle | Elongation |
| --> 20 cycles | ||
| 68 | 600 | Final elongation |
| 6 | Hold |
Method:
- pipetting the mastermix on ice
- preheating of the thermocycler: starting the program IGMDN1 (Eppendorf Mastercycler gradient)
- finally adding polymerase
- start the PCR program (press enter)
Results:
- PCR product
Further tasks:
- Verification of the PCR product using agarose gel electrophoresis
PCR of mdnA
Investigators: Jessica, Nicole
Aim: Testing the possibility of amplifying mdnA using PCR and the chosen conditions
Time: 2011-06-08,9:30-14:00
Materials:
- DNA polymerases: NEB OneTaq DNA Polymerase
- Template DNA: pARW089 (vector)
- Forward primer: sf_mdna_1
- Reverse Primer: r_mdna_1
--> mastermix composition, see 11th labday 'Design of PCR conditions for amplifying mdnA'
- Eppendorf Mastercycler personal
- used temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 60 | Initial denaturation |
| 94 | 20 | Denaturation |
| 56 | 20 | Annealing |
| 68 | 20 | Elongation |
| --> 30 cycles | ||
| 68 | 300 | Final elongation |
| 6 | Hold |
Method:
- pipetting the mastermix on ice
- preheating of the thermocycler: starting the program IGMDNA1 (Eppendorf Mastercycler personal)
- finally adding polymerase
- start the PCR program (press enter)
Results:
- PCR product
Further tasks:
- Verification of the PCR product using agarose gel electrophoresis
Production of agarose gel
Investigators: Jessica, Nicole
Aim: Verification of PCR product, mdnA and whole mdn cluster, by agarose gel electrophoresis
Time: 2011-06-08,9:30-14:00
Materials:
- Agarose, type VII, low gelling temperature (Sigma)
- 1x TAE Buffer (produced by AG Biotechnology)
- GelRed
Method:
1. mdn cluster (approx. 6712 bp) --> 0.8 %
- 0.75 g agarose in 50 ml 1xTAE buffer
- dissolving/ boiling up using microwave
- after cooling, adding of 2 µl GelRed
2. mdnA (approx. 276 bp) --> 1.5 %
- 0.4 g agarose in 50 ml 1xTAE buffer
- dissolving/ boiling up using microwave
- after cooling, adding of 2 µl Gelred
Results
- 1.5 % agarose gel
- 0.8 % agarose gel
Further tasks:
- Loading gel and performing electrophoresis
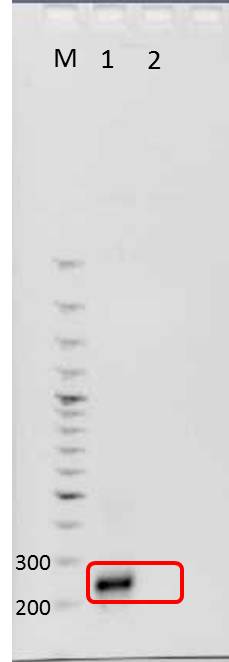
Agarose gel electrophoresis of mdnA
Investigators: Jessica, Nicole, Nadja
Aim: Verification of PCR product, mdnA, by agarose gel electrophoresis
Time: 2011-06-08,15:45-18:00
Materials:
- 1.5 % agarose gel, produced previously
- PCR product of mdnA amplification
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Loading samples
- dilution of PCR product: 1 µl DNA and 4 µl water
- Adding 1 µl 6x loading dye to diluted PCR product
Positions of samples/ marker
| lane | Sample | Sample/[µl] | Expected size (bp) | approx. size | |
|---|---|---|---|---|---|
| 1 | marker | 2 | |||
| 2 | PCR product mdnA | 6 | 276 | 280 | |
| 4 | PCR product mdnA (dilution) | 6 | 276 | 280 | |
| 5 | marker (dilution 1:5) | 2 |
2. Run
- 100 V
- time: 1 h
Results:
approx. size: see methods
Design of PCR conditions for amplifying mdnA for Phage display
Investigators: Sandrina, Sabine
Aim: PCR reaction of mdnA with overlapping primers containing SfiI restriction sites for cloning into PAK100, which is needed for phage display
Time: 2011-06-8,12:00-14:00
Materials:
- Sequence of vector pARW089
- Manual: OneTaq Polymerase (NEB)
- Forward primer: pf_mdnA_sfi_1
- Reverse primer: pr_mdnA_sfi_1
- Geneious
Method:
- Design of a temperature profile based on the manual
Results:
1. Temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 60 | Initial denaturation |
| 94 | 20 | Denaturation |
| 60 | 20 | Annealing |
| 72 | 20 | Elongation |
| --> 15 cycles | ||
| 94 | 60 | Denaturation |
| 70 | 40 | Annealing |
| 72 | 20 | Elongation |
| --> 15 cycles | ||
| 70 | 300 | Final extension |
Further tasks:
PCR
13th Labday 2011-06-09
Agarose gel electrophoresis of mdn cluster
Investigators: Jessica, Steffi, Nicole
Aim: Verification of amplification of the complete mdn cluster by PCR
Time: 2011-06-09,10:00-17:30
Materials:
- 0.8 % agarose gel, produced previously
- PCR product of mdn amplification
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Loading samples
- dilution of PCR product:
- Adding 1 µl 6x loading dye to diluted PCR product
Positions of samples/ marker
| lane | Sample | Sample/[µl] | Expected size (bp) | approx. size | |
|---|---|---|---|---|---|
| 1 | marker | ||||
| 2 | PCR product mdn cluster | ||||
| 4 | PCR product mdn cluster (dilution) | ||||
| 5 | PCR product mdn cluster (dilution) |
2. Run
- 100 V
- time: 1 h
Results:
approx. size: see methods
14th Labday 2011-06-10
Amplification of mdnA for phage display
Investigators: Sandrina, Sabine
Aim: amplificate mdnA of pARW089 with primers pf_mdnA_sfi_1 and pr_mdnA_sfi_1
Time: 2011-06-10,08:00-10:00, 14:00-20:00
Materials/Methods:
1. PCR (see 11th Labday 2011-06-08)
- 0,75 ng Vector pARW089
- Thermo Hybraid Px2
- 0,25 µl OneTaq DNA Polymerase (NEB)
- 1 µl dNTPs
- 1 µl each forward and reverse primer
- 32,15 µl DNase free water
- 10 µl 5xOneTaq Standard Reaction Buffer
2. PCR product purification
- QIAquick Gel Extraction Kit (250)
3. Digestion with SfiI
- 50 µl sample
- NEB 10x buffer 4
- 100x BSA
- 3 µl restriction enzyme SfiI
- PCR product (mdnA with SfiI restriction sites)
Further tasks:
gel electrophoresis and gel extraction
15th Labday 2011-06-14
cultivation of cells containing pAK100 from E. coli glycerol stocks
Investigators: Sandrina
Aim: cultivate cells conatining pAK100 (vector) to purify it and use it for phage display.
1.amplificated mdnA should be cloned into pAK100
2. genIII should be amplified from it to clone into pARW089
Time: 2011-06-14,13:45-14:45
Materials/Methods:
- glycerol stock nr. 15, pAK100 bla KDIR, XL1-blu, 2003-07-31
- LB-medium with chloramphenicol (1:1000)
Further tasks:
plasmid preperation und digestion with SfiI
16th Labday 2011-06-15
gel electroporesis PCR product(mdnA, 2011-06-10), plasmid preperation und digestion with SfiI of pAK100 bla KDIR
Investigators: Sandrina, Sabine, Jessica
Aim: purification of mdnA and pak100 to use it for phage display.
1.amplificated mdnA should be cloned into pAK100
2.genIII should be amplified from it to clone into pARW089
Time: 2011-06-15,12:00-18:00
Materials/Methods:
1. plasmid preparation
GeneJET Plasmid Miniprep Kit (Fermentas, K0509)
2. Digestion with SfiI
- 50 µl sample
- NEB 10x buffer 4
- 100x BSA
- 3 µl restriction enzyme SfiI
- PCR product (mdnA with SfiI restriction sites)
3. gel electrophoresis
sample preparation: 50 µl PCR product (digested) + 10 µl 6x loading dye solution
0,75 g Agarose, 50 ml TAE (1 x), 2 µl GELRED, at 100 Volt,running time: 45 minutes
marker: GeneRuler ladder 100 Bp plus ladder (Fermentas)
Results:
no results, PCR didn`t work
Further tasks:
repeat PCR, gel electrophoresis of digested pak100
17th Labday 2011-06-16
gel electrophoresis and gel extraction of digested pAK100 bla KDIR
Investigators: Sandrina, Sabine, Anna
Time: 2011-06-16,10:00-12:00
Aim: get a pAK100 plasmid digested with SfiI to clone mdnA into it (phage display, strategy 1)
Materials/Methods:
- 1.gel electrophoresis sample preparation: 50 µl plasmid product (digested) + 10 µl 6x loading dye solution 0,5 g Agarose, 50 ml TAE (1 x), 2 µl GELRED, at 100 Volt,running time: 45 minutes marker: 1 kb DNA ladder (NEB)
- 2. gel extraction
QIAquick Gel Extraction Kit (250)
Results:
digestion worked, plasmids were extracted from the gel and stored at -20 °C
Further tasks:
ligation of digested pAK100 with amplified mdnA containing SfiI restriction sites
PCR of mdnA for phage display repeated with modified conditions
Investigators: Sandrina, Sabine, Anna
Time: 2011-06-16,10:00-12:00
Aim: amplification of mdnA with SfiI restriction sites (vector pARW089)
Materials/Methods:
see 2011-06-10
changes:
- annealing temperature 56°C (stage 1)
- template 65 ng
gel electrophoresis of PCR product
Investigators: Sandrina, Anna, Sabine
Time: 2011-06-16,16:00-18:00
Material and Methods:
see 2011-06-15
Results:
no result
reason: reversed primer was ordered unreversed
Further tasks:
order a new primer
Over night culture of E. coli XL1-blue and RV308
Investigators: Sandrina, Anna, Sabine
Time: 2011-06-16,18:00-19:00
Aim: cultering cells to produce competent cells
Material and Methods:
- XL1-blue: 15 ml LB medium with 100 µg/mL tetracycline out of tetracycline stock solution 50 mg/mL
- RV308: 15 ml LB medium without streptomycin (not available)
Further tasks:
produce competent cells
18th Labday 2011-06-17
competent cells - E. coli XL1 blue & RV308
Investigator: Niels
Aim: produce competent cells
Time: 2011-06-15,09:00-16:30
Materials/Methods:
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- Prepare 45 Eppis (1,5µl)
- get liquid nitrogen
- Use Milipore-filter for sterile CaCl2 , keep cool!
- prepare 15ml LB-Medium (or DYT) with the specific antibiotic (XL1-blue Tet, BL21 none!), inoculate and incubate over night
- prepare 100ml LB-Medium (or DYT) with the specific antibiotic, inoculate with 2ml of the over-night-culture. Nurture the culture until OD600 at 0,35 (0,2-0,5) (if the OD is too high, the cell won’t be competent)
- keep cell suspension in sterile falcons (50ml) 20 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend on ice with 10ml cold CaCl2-solution (put a little of the 10ml solution in every falcon before!), pool every resuspended aliquot of one cell line and add 40ml CaCl2-solution (total volume 50ml). Keep 30 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend pellet in 5,5ml CaCl2(80mM)/Glycerol (4:1), use the pippete boy and a sterile glas pipette to dissolve the pellet
- aliquot in Eppis á 150µl and store immediately at liquid nitrogen and afterwards at -80 °C
Results:
- 22 tubes RV308 for expression(150µl competent cells at -80°C)
- 22 tubes XL1 blue for transformation(150µl competent cells at -80°C)
Further tasks:
check by transformation
primer design for phage display
Investigators: Jessica, Sabine, Sandrina, Anna
Aim: Design primers for PCR of
- genIII (from pak100)
- mdnA (from pARW089)
to fuse genIII and mdnA in pARW089
Time: 2011-06-17,14:00-18:00
Materials:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
Primer design
- melting temperature based on 4+2 method (for mdnA-overlapping part)
- adding sequences for restriction sites (considering reading frame)
- Check self-complementarity and melting temperature using Oligo Calc
Results:
Primer sequences (strategy 2)
- pf_iGEM_GenIII_NgoMIV TAAGCTGCCGGCGAGCAGAAGCTGATCTCTGAGGAAGACCTGGGTGGTGGCTCTGGTTCC (2nt-NgoMIV-myc-GenIII)
- pr_GenIII_iGEM_AatII 5'->3' TGCTTAGACGTCTACTAGTATTAACCGGTAGACTCCTTATTACGCAGTA (GenIII-AgeI-SpeI-AatII-6nt)
- pf_mdnA-iGEM_EheI TTCCATGGCGCCAGAGGAATCTAGATGGCATATCCCAACGATC (6nt-EheI-RBS-XbaI-mdnA)
- pr_mdnA_iGEM_AAtII 5'->3' GACGTCCTGCAGCGGCCGCTACTAGTATTAACCGGTTATAATCTTCCCAGTCAGAAG (mdnA-AgeI-SpeI-NotI-PstI-AatII-nt)
new ordered primer for amplification of mdnA from pARW089 with SfiI restriction sites (strategy 1)
- pr_sfi_mdna_1 5'->3' AGCCTGCAGCGGCCGCTACTAGTATTAGTGGCCCCCGAGGCCATAATCTTCCCAGTCAGAAGGG
Further tasks:
PCR
19th Labday 2011-06-20
Preparation of LB-Agarplates
Investigators: Nadine, Katharina, Steffi, Nicole, Jessica
Time: 2011-06-09,13:45-19:00
Aim: Preparation of LB-Agarplates
Recipe:
- 10 g/l Yeast-Tryptone
- 10 g/l NaCl
- 5 g/l Yeast-Extract
- 15 g Agar-Agar
- fill up to 1000 ml with Millipore H2O and autoclave
Further tasks:
add antibiotics (Ampicillin, Kanamycin) to LB-Medium and prepare plates
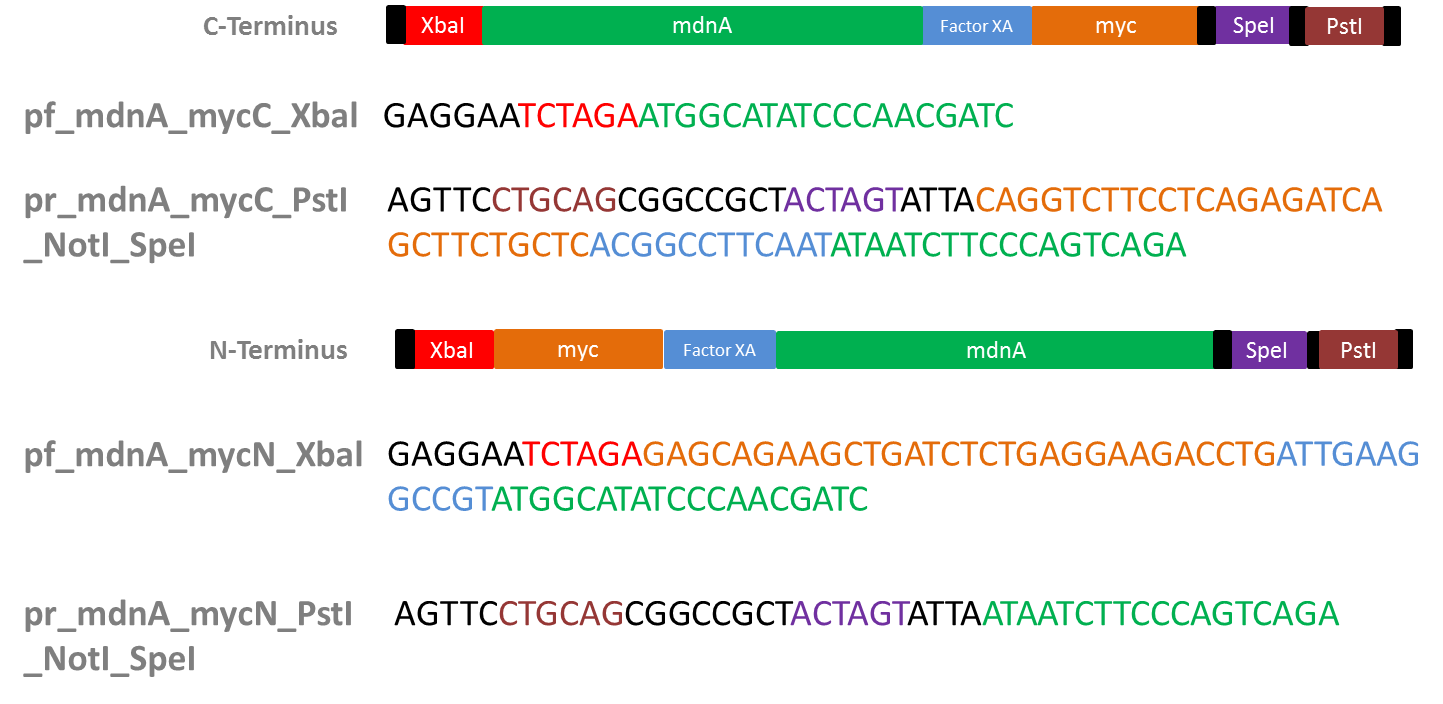
Design of Primers for myc-tagging mdnA
Investigators: Nicole, Nadine, Katharina, Steffi
Aim: Design primers for mdnA (from pARW089) in order to tag it w/ myc (+ Factor XA Protease restriction site) to clone into vector from Sven (iGEM vector from 2010)
Time: 2011-06-01,16:00-19:00
Materials:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
- Primer design
- melting temperature based on 4+2 method (for mdnA-overlapping part)
- adding sequences for restriction sites (considering reading frame)
- Check self-complementarity and melting temperature using Oligo Calc
- Myc-tag
- why?: purification by affinity chromatography (using antybodies against myc-tag)
- Factor XA Protease
- why?: to be able to cleave myc tag after mdnA expression and purification
- restriction site: I E/D G R
- used sequence (forward): ATT GAA GGC CGT
Results:
Primer sequences
Further tasks:
- do PCR of mdnA
20st Labday 2011-06-21
Error prone PCR (varied MnCl2 and MgCl2 concentration) of mdnA
Investigators: Nadine, Steffi, Nicole
Time: 2011-06-21,9:00-14:00
Aim: Random mutation of mdnA gene based on increased MnCl2 and MgCl2 concentration
Materials:
- Marcus Schicklberger's (trainee, August 2005) protocol performing several error prone PCRs
- Manual Supplement of Cadwell and Joyce 2009 [[Media:
- Genaxxon Bioscience Taq S Polymerase, manufacturer's protocol Media: UP_Genaxxon_Taq_Polymerase_Protocol.pdf
- Genaxxon Bioscience taq Polymerase
- 10x Genaxxon Bioscience taq Polymerase buffer containing 15 mM MgCl2
- dNTPs (10 mM)
- forward primer: sf_mdnA_1 (10 µM)
- reverse primer: r_mdnA_1 (10 µM)
- template DNA pARW089 (857,3 ng/ µl measured by nanotrop a second time)
- Genaxxon Bioscience MgCl2 stock solution (50 mM)
- Manganese(II) chloride (Merck)
- MnCl2
- sterile filter
- Eppendorf Mastercycler personal, program IGMDNA1
Method:
Production of 50 mM Manganese(II) chloride stock solution
- MMnCl2 = 161.81 g/ mol
- 50 mM in 1 ml needed --> mMnCl2 = 8.1 mg
- sterile filtration: pore size of filter: 0.2 µm
Error prone PCR of mdnA
- increasing MnCl2 concentration beginning with 0 mM in 0.05 mM steps, ending with 0.15 mM (--> sample 1-4)
- increasing MgCl2 concentration beginning with 1.5 mM, in 5 mM steps, ending with 15 mM (--> sample 5-8)
1. Mix for error prone PCR with increasing MnCl2 concentration
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| Genaxxon Bioscience Taq Polymerase buffer containing MgCl2) | 15 mM (MgCl2) | 5 | 1.5 mM MgCL2 |
| MgCl2 | 50 mM | 5.5 | 5.5 mM |
| dNTPs | 10 mM | 2.0 | 0.4 mM |
| Primer (each) | 10 µM | 2.5 | 0.5 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 5.58 | 0.7 ng |
| Polymerase | 1 | 5 U |
in addition MnCl2 and H2O as shown following
| Sample number | MnCl2 in µl | Final concentration MnCl2 | H2O in µl |
|---|---|---|---|
| 1 | 0 | 0 | 25.52 |
| 2 | 0.5 | 0.05 mM | 23.02 |
| 3 | 1.0 | 0.1 mM | 22.52 |
| 4 | 1.5 | 0.15 mM | 22.02 |
2. Mix for error prone PCR with increasing MgCl2 concentration
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| Genaxxon Bioscience Taq Polymerase buffer containing MgCl2) | 15 mM (MgCl2) | 5 | 1.5 mM MgCL2 |
| dNTPs | 10 mM | 2.0 | 0.4 mM |
| Primer (each) | 10 µM | 2.5 | 0.5 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 8.0 | 5 ng |
| Polymerase | 1 | 5 U |
in addition MgCl2 and H2O as shown following
| Sample number | MnCl2 in µl | Final concentration MnCl2 | H2O in µl |
|---|---|---|---|
| 5 | 0 | 1.5 mM | 29.0 µl |
| 6 | 3.5 | 5 mM | 17.5 µl |
| 7 | 7.0 | 10 mM | 14.0 µl |
| 8 | 10.0 | 15 mM | 10.5 µl |
3. Complete pipet scheme
| Component in µl | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Template DNA | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| dNTPs | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 10x Genaxxon polymerase buffer (with MgCl2) | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| MgCl2 (50 mM) | 5.5 | 5.5 | 5.5 | 5.5 | 0 | 3.5 | 7.0 | 10.5 |
| MnCl2 (5 mM) | 0 | 0.5 | 1 | 1.5 | 0 | 0 | 0 | 0 |
| Forward primer (sf_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Reverse primer (r_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| H2O | 23.5 | 23.0 | 22.5 | 22.0 | 29.0 | 25.5 | 22.0 | 18.5 |
4. Temperature profile
- see 12th labday
- Eppendorf Mastercycler personal, program IGMDNA1
Results:
- 8 PCR products containing mutated mdnA
Further tasks:
- Verification of these PCR products by agarose gel electrophoresis
- Purification and cloning of positive samples
- Sequencing
Agarose gel electrophoresis of mutated mdnA
Investigators: Nadine, Jessica, Steffi, Nicole
Time: 2011-06-21,14:00-17:00
Aim: Verification of random mutated mdnA (mutations are based on increased MnCl2 and MgCl2 concentration)
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- 8 PCR products of mutated mdnA, see
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of two 1 % agarose gel
For each gel:
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 30 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler
Positions of marker and sample
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | mdnA 1 | 36 | 276 |
| 3 | mdnA 2 | 36 | 276 |
| 4 | mdnA 3 | 36 | 276 |
| 5 | mdnA 4 | 36 | 276 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | mdnA 5 | 36 | 276 |
| 3 | mdnA 6 | 36 | 276 |
| 4 | mdnA 7 | 36 | 276 |
| 5 | mdnA 8 | 36 | 276 |
3. Run
- 100 V
- time: 1 h
Results:
- Gel Extraction DNA excision: lane 1-4, 5,7: ~ 276 bp band
- stored at -20°C in orange box (iGEM 2011 UP DNA)
Further tasks:
- DNA extraction
- Cloning
- Sequencing
Over night culture of E. coli BL21 containing BBa_K404304_pSB1C3 (biobrick iGEM Freiburg 2010) from Sven
Investigators: Jessica, Nadine, Stefan, Steffi
Aim: Culturing E. coli containing BBa_K404304_pSB1C3 biobrick for midi-prep and using for cloning afterwards
Time: 2011-06-21,16:50-17:00
Materials:
- cells: E. coli BL21 containing BBa_K404304_pSB1C3 (biobrick iGEM Freiburg 2010) from Sven
- 50 ml LB media
- 50 µl Chloramphenicol (100 mg/ml)
Method:
- add antibiotic to LB
- inoculation of cells
- 37 °C, 250 rpm, 17 hours
Results:
Further tasks:
- remove cells from shaker at 10:00 (Nici :D)
- do midi-prep
Transformation of E. coli XL 1 blu and RV 308 + puK
Investigators: Niels
Aim: Check the quality of competent cells
Time: 2011-06-21,10:00-12:30
Materials:
- cells: E. coli XL 1 blu and RV 308
- 50 ml LB media
- 50 µl ampicillin (100 mg/ml)
Method:
protocol: addition of 1 µl plasmid (puK) to cells in 1.5 ml Eppi,
- incubation 30 min on ice,
- heat shock 90 sec at 42°C,
- addition of 750 µl LB medium,
- incubation at 37 °C for 60 min in Eppendorf thermomixer at 450 rpm,
- centrifugation at 2000xg; for 5 min,
- decandation of supernatant,
- resuspension of pellet in approx. 100 µl,
- plating on LB medium with 1,5 % agar and LB medium with 1,5 % agar, 100 µg/ml ampicillin,
- storage over night at 37°C
Results:
- negative
Further tasks:
- repeat
21th Labday 2011-06-22
Gel extraction of mutated mdnA
Investigators: Katharina, Nicole
Aim: Extraction of mdnA, which was amplified and mutated (using error prone PCR)
Time: 2011-06-22,12:15-13:30
Materials:
- Qiagen QIAquick Gel extraction Kit
- PCR products 1-5 and 7 containing mutated mdnA
Method:
- performing gel extraction based on manufacturer's protocol, Media: UP_Qiagen_QIAquick_Gel_Extraction_kit.pdf
- elute in 30 µl buffer EB
Results:
- purified PCR products of mutated mdnA (1-5, 7)
Further tasks:
- restriction enzyme digestion with purified mdnA and pARW089 using AatII/ NarI
- Verification by agarose gel electrophoresis
- Ligation
Midiprep of pSB1C3 carrying cells
Investigators: Nadine, Vanessa
Aim:
- Midiprep of pSB1C3
- vector for biobricks (RFC25)
Time: 2011-06-22, 14:00-16:00
Materials:
- Qiagen Plasmid Plus Midi Kit
- over night cultures: E. coli BL21 containing BBa_K404304_pSB1C3 (biobrick iGEM Freiburg 2010) from Sven
- NanoDrop
Method:
Midiprep
- using Qiagen Plasmid Plus Midi Kit Quick-Start Protocol, Media: UP_QIAGEN_Plasmid_Plus_Midi_Kit.pdf
Output: 4 x DNA Eppi Tubes: pSB1C3, 22.06.2011, storage: orange box (iGEM DNA, -20°C)
Results:
- concentration:
- tube 1: 284.6 ng/µl, 260/280 = 1.90, 260/230 = 2.32
- tube 2: 252.6 ng/µl, 260/280 = 1.91, 260/230 = 2.34
- tube 3: 235.0 ng/µl, 260/280 = 1.89, 260/230 = 2.01
- tube 4: 238.4 ng/µl, 260/280 = 1.9, 260/230 = 2.32
Further tasks:
Restriction enzyme digestion and agarose gel electrophoresis of mutated mdnA
Investigators: Katharina, Nicole
Time: 2011-06-22,14:00-18:00
Aim: Restriction enzyme digestion of random mutated mdnA and pARW089 (vector) and verification using gel electrophoresis
Materials:
- Restriction enzymes: AatII, NarI
- NEB Buffer 4
- PCR products of random mutated mdnA(1-5, 7), purified from gel
- vector pARW089
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
- 1kb DNA ladder (NEB)
Method:
1. Restriction enzyme digestion
| components | volume of PCR products /µl | volume of pARW089 /µl |
| DNA | 30 | 6 |
| NEB Buffer 4(10x) | 4 | 1 |
| Enzyme AatII | 1 | 1 |
| Enzyme NarI | 1 | 1 |
| H2O | 4 | 1 |
| Total volume | 40 | 10 |
2. Production of two 1 % agarose gels
- mAgarose = 1.0 g in 100 ml 1x TAE
- adding 4 µl gel red
3. Loading samples
a. restriction enzyme digestion products of random mutated mdnA
- 30 µl PCR product and 6 µl 6x loading dye
b. restriction enzyme digestion product of vector pARW089
- 10 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler, 100bp plus (Fermentas)
- 2 µl 1 kb DNA ladder (NEB)
Positions of marker and sample
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | mdnA 1 | 36 | |
| 3 | mdnA 2 | 36 | |
| 4 | mdnA 3 | 36 | |
| 5 | mdnA 4 | 36 | |
| 6 | mdnA 5 | 36 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | mdnA 7 | 36 | |
| 5 | pARW089 | ||
| 6 | marker, 1kb DNA ladder (NEB) | 2 |
4. Run
- 100 V
- time: 1 - 1.5 h
Results:
- stored in freezer
Further tasks:
- DNA extraction
- Cloning
- Sequencing
Over night culture of E. coli containing pEX respectively pBAD from Tobias
Investigators: Jessica, Nadine, Nicole
Aim: Culturing E. coli containing PEX respectively pBAD for midi-prep
Time: 2011-06-22,16:50-17:00
Materials:
- cells: E. coli containing pEX respectively pBAD from Tobias
- 50 ml LB media
- 50 µl Ampicillin (100? mg/ml)
Method:
- add antibiotic to LB
- inoculation of cells
- 37 °C, 250 rpm, 17 hours
Results:
Further tasks:
- remove cells from shaker at 10:00
- do midi-prep
22th Labday 2011-06-23
Midiprep of pEX respectively pBAD carrying cells
Investigators: Jessica
Aim:
- Midiprep of pEX respectively pBAD
- expression vectors
Time: 2011-06-23, 10:30-12:30
Materials:
- Qiagen Plasmid Plus Midi Kit
- over night cultures: E. coli containing pEX respectively pBAD
- NanoDrop
Method:
Midiprep
- using Qiagen Plasmid Plus Midi Kit Quick-Start Protocol, Media: UP_QIAGEN_Plasmid_Plus_Midi_Kit.pdf
Results:
- concentration:
- pEX tube 1: 36.4 ng/µl
- pEX tube 2: 62.4 ng/µl
- pBAD tube 1: 71.6 ng/µl
- pBAD tube 2: 94.6 ng/µl
Output:
- 2 x DNA Eppi Tubes: pBAD, 23.06.2011, storage: orange box (iGEM DNA, -20°C)
- concentration may be okay as pBAD is a low-copy plasmid
- pEX tubes thrown away because of low concentration
Further tasks:
- redo culturing and midiprep for pEX
Over night culture of E. coli XL1-Blue and RV308 for competent cells
Investigators: Stefan
Aim: Culturing cells to produce competent cells
Time: 2011-06-23,15:30-16:00
Materials:
- XL1-Blue: 15 ml LB medium with 100 µg/mL tetracycline out of tetracycline stock solution 50 mg/mL
- RV308: 15 ml LB medium without streptomycin (not available)
Method:
- add antibiotic to LB
- inoculation of cells
- 30 °C, 250 rpm, 18 hours
- 37 °C, 250 rpm, 5 hours
- inoculate 200 mL fresh culture with 2 mL from starter culture
Results:
Further tasks:
- produce competent cells
Over night culture of E. coli containing vector pBAD resp. pEX for midiprep resp. glycerol stocks
Investigators: Nicole
Aim: Culturing cells to do midiprep and to produce glycerol stocks
Time: 2011-06-23,17:30-18:00
Materials:
- E. coli containing expression vector pBAD: 5 ml LB medium with 5 µl ampicilin
- E. coli containing expression vector pEX: 50 ml LB medium with 50 µl ampicilin
Method:
- add antibiotic to LB
- inoculation of cells
- 37 °C, over night
Results:
Over night culture of
- E. coli containing expression vector pBAD
- E. coli containing expression vector pEX
Further tasks:
- Midiprep for E. coli containg vector pEX
- glycerol stocks of E. coli containg vector pEX as well as fpr E. coli containg vector pBAD
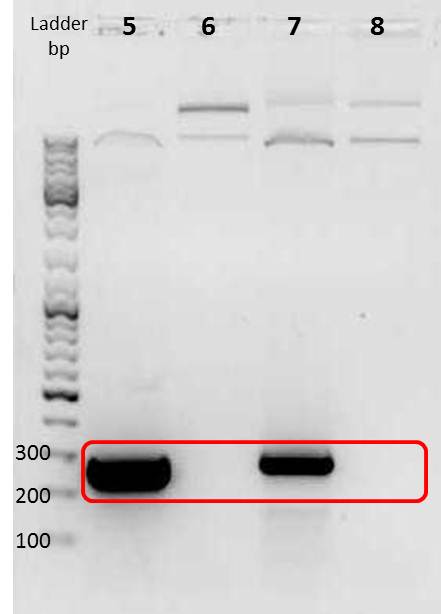
Amplification of mdnA and GeneIII for phage display
Investigators: Sandrina, Sabine
Aim:
amplificate mdnA of pARW089 with primers pf_mdnA_sfi_1 and pr_mdnA_sfi_1 (phage display strategy 1)
amplificate mdnA of pARW089 with primers pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (phage display strategy 2)
amplificate GeneIII of pak100 with primers pf_iGEM_GenIII_NgoMIV and pr_GenIII_iGEM_AatII (phage display strategy 2)
Time: 2011-06-23, 10:00-12:00, 15:00-17:00
Materials/Methods:
1. PCR
- 0,8 ng Vector pARW089
- Thermo Hybraid Px2 (conditions see 2011-06-08, changes: extension time 40 s)
- 0,25 µl OneTaq DNA Polymerase (NEB)
- 1 µl dNTPs
- 1 µl per primer
- 31,75 µl DNase free water
- 10 µl 5xOneTaq Standard Reaction Buffer
2. gel electrophoresis (analytic)
- 5 µl PCR product, agarose gel: 1%)
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
Positions of marker and sample
Gel
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker | 2 | |
| 1 | mdnA with SfiI | 7 | 224 |
| 2 | mdnA iGEM | 7 | ca. 230 |
| 3 | geneIII iGEM | 7 | ca. 520 |
Results:
File:UP PCR sfi mdna,mdna rfc25,genIII.png
PCR for strategy 1 (lane number 1) probably did not work because the annealing temperature of the forward primer is located at 42°C (not 60 °C)
Furter tasks:
- purification of PCR products
- order new forward primer for strategy 1 with higher annealing temperature
Design of primers and planing of further experiments
Investigators: Nadine, Nicole
Time: 2011-06-23, 15:00-18:00
Aim: Design of primers and detailed planing of further experiment
Materials:
1. Primer design
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
1. Primer Design
- add an start codon (ATG) to the previously designed primer pf_mdnA_mycN_xbal
2. Detailed planing of further experiments
A. Control experiment of error prone PCR of mdnA and subsequent restriction enzyme digestion using NarI and AatII
Aim: Analysis of three lines after agarose gel electrophoresis after restriction enzyme digestion of mdnA using NarI and AatII; see
Idea: DNA is not completly single stranded; would be avoid by heating mixture after restriction enzyme digestion for 10-20 min at 65°C
B. Random mutagenesis of mdnA and determination of mutation rate
Idea: using an increased MnCl2 concentration to mutate mdnA and finally sequencing of mdnA to determine the mutation rate
C. mdnA as iGEM expression part in vector pEX or pBAD
D. mdnA as biobrick
Idea: Cloning of mdnA in iGEM cloning vector to submit as biobrick
E. Repetition of Elke's experiments as control for further experiments
Idea: Further, purify mdnA with the use of antibodies (anti myc-tag) and without HPLC. To compare which strategy is more efficient (myc-tag or HPLC) HPLC experiment as control is necessary.
F. mdnA with myc-tag (N- and C-terminal) in iGEM expression vector pEX or pBAD
Idea: myc-tagging of mdnA to purify it with antibodies (without use of HPLC)
G. Expression and purification of mdnA (with myc-tag)
Idea: Determine the efficiency of myc-tag/ antibody to purify mdnA.
| To do | Experimental tasks |
|---|---|
| A. | Control experiment for error prone PCR (increased MnCl2 concentration) of mdnA (vector pARW089) |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of (positive) samples | |
| Restriction enzyme digestion of mdnA using NarI and AatII; Splitting the samples fifty-fifty, heat one half of each sample 10-20 min at 65°C | |
| Analytic agarose gel electrophoresis of each sample (heated and non-heated) | |
| B. | Random mutagenesis of mdnA and determination of mutation rate |
| Error prone PCR of mdnA (in vector pARW089) using increased MnCl2 concentration | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the positive samples | |
| Restriction enzyme digestion of mdnA and pARW089 using NarI and AatII | |
| Preparative agarose gel electrophoresis of each sample | |
| DNA extraction and purification of the positive samples | |
| Ligation | |
| Transformation | |
| DNA isolation of one clone using midiprep/ miniprep | |
| Sequencing | |
| C. | mdnA as expression part |
| PCR of mdnA in pARW089 using pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (primer designed by phage display group) | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the positive sample | |
| Restriction enzyme digestion of mdnA and iGEM expression vector pEX or pBAD | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the samples | |
| Ligation | |
| Transformation | |
| DNA isolation of one clone using midiprep/ miniprep | |
| Sequencing | |
| D. | mdnA in cloning vector |
| PCR of mdnA in pARW089 using pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (primer designed by phage display group) | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the positive sample | |
| Restriction enzyme digestion of mdnA and iGEM expression vector pEX or pBAD | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the samples | |
| Ligation | |
| Transformation | |
| DNA isolation of one clone using midiprep/ miniprep | |
| Sequencing | |
| E. | Repetition of Elke's experiments as control for further experiments |
| Expression and purification (HPLC; without myc-tag) of mdnA x mdnA in pARW089 x mdnA in iGEM expression vector pEX or pBAD | |
| F. | mdnA with myc-tag (N- and C-terminal) in iGEM expression vector pEX or pBAD |
| PCR of mdnA in pARW089 using x C-terminal: pf_mdnA_mycC_Xbal (forward) and pr_mdnA_mycC_PstI_NotI_SpeI (reverse) x N-terminal: pf_mdnA_mycN_Xbal_1 (forward) and pr_mdnA_mycN_PstI_NotI_SpeI (reverse) | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the positive samples | |
| Restriction enzyme digestion of mdnA and iGEM expression vector pEX or pBAD | |
| Preparative agarose gel electrophoresis | |
| DNA extraction and purification of the samples | |
| Ligation | |
| G. | Expression and purification of mdnA (with myc-tag) x N-terminal myc-tag x C-terminal myc-tag |
| Transformation of E. coli with pEX/ pBAD containing mdnA with myc-tag and with pARW089 |
23th Labday 2011-06-24
purification of PCR products 2011-06-23
Investigators: Sandrina
Aim:
- purification of PCR products mdnA iGEM and geneIII iGEM (lanes number 2 + 3) for phage display (strategy 2)
Time: 2011-06-24, 14:30-15:30
Materials/Methods:
- QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion of the two fragments with EheI and AgeI (mdnA iGem) and AatII and NgoMIV (geneIII iGEM)
- digestion of vector pARW089 with EheI and AatII
preparation of competent E. coli XL1 and RV 308
Investigators: Stefan
Aim:
- get competent cells of E. coli XL 1 blue and RV 308
Time: 2011-06-24, 14:00-19:30
Materials:
- over night cultures: E. coli XL 1 blu and RV 308
Method:
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- The cooling-centrifuge is in the tool shed, cool down to 4°C early enough, close the lid correctly!
- Prepare early enough min. 100 Eppis (1,5µl) (per cellline) and cool down to -80°C before using
- Use Milipore-filter for sterile CaCl2 , keep cool!
- prepare 15ml LB-Medium (or DYT) with the specific antibiotic (XL1-blue Tet, BL21 none!), inoculate and incubate over night
- prepare 200ml LB-Medium (or DYT) with the specific antibiotic, inoculate with 2ml of the over-night-culture. Nurture the culture until OD600 at 0,35 (0,2-0,5) (if the OD is too high, the cell won’t be competent)
- keep cell suspension in sterile falcons (50ml) 20 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend on ice with 10ml cold CaCl2-solution (put a little of the 10ml solution in every falcon before!), pool every resuspended aliquot of one cell line and add 40ml CaCl2-solution (total volume 50ml). Keep 30 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend pellet in 5,5ml CaCl2(80mM)/Glycerol (4:1), use the pippete boy and a sterile glas pipette to dissolve the pellet
- aliquot in Eppis á 60µl and store immediately at - 80 °C
Results:
- 91 aliquots a 60 µL of XL1 and RF
Output:
Further tasks:
test the cells
Eppis in - 80°C in Boxen geräumt, aber noch nicht umbeschriftet
Midiprep of pEX carrying cells
Investigators: Nadine
Aim:
- Midiprep of pEX
Time: 2011-06-24, 10:00-12:00
Materials:
- Qiagen Plasmid Plus Midi Kit
- over night cultures: 50 ml E. coli containing pEX from Tobi
- NanoDrop
Method:
Midiprep
- using Qiagen Plasmid Plus Midi Kit Quick-Start Protocol, Media: UP_QIAGEN_Plasmid_Plus_Midi_Kit.pdf
Output: 1x DNA Eppi Tubes: #5 pEX, 24.06.2011, storage: orange box (iGEM DNA, -20°C)
Results:
- concentration:
- tube 1: 248.4 ng/µl
Further tasks:
Glycerol stocks of E. coli cells carrying pEX and pBAD plasmids
Investigators: Nadine
Aim: Generating stocks for further research
Time: 2011-06-24,10:00-10:15
Materials:
- Glycerol
- 5 ml over-night culter of E. coli pBAD
- 50 ml over-night culter of E. coli pEX (also for midi-prep)
Method:
- 700 µl of cell suspension
- 300 µl glycerol
- mixed gently and frozen by -80°C
24th Labday 2011-06-27
Control experiment fpr error prone PCR and enzyme digestion of mdnA
Investigators: Nadine, Steffi, Katharina, Nicole
Time: 2011-06-27, 14:30-16:30
Aim: control experiment to verify error prone PCR and enzyme digestion of mdnA, which experiment was done previously
--> error prone PCR using increased MnCl2 concentration
Materials:
- protocol of experiment, which was done previously; see2011-06-22
- Genaxxon Bioscience taq Polymerase
- 10x Genaxxon Bioscience taq Polymerase buffer containing 15 mM MgCl2
- dNTPs (10 mM)
- forward primer: sf_mdnA_1 (10 µM)
- reverse primer: r_mdnA_1 (10 µM)
- template DNA pARW089 (857,3 ng/ µl)
- Genaxxon Bioscience MgCl2 stock solution (50 mM)
- MnCl2
- MnCl2 stock solution (50 mM)
Method:
1. Mix for error prone PCR with increasing MnCl2 concentration
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| Genaxxon Bioscience Taq Polymerase buffer containing MgCl2) | 15 mM (MgCl2) | 5 | 1.5 mM MgCL2 |
| MgCl2 | 50 mM | 5.5 | 5.5 mM |
| dNTPs | 10 mM | 2.0 | 0.4 mM |
| Primer (sf_mdnA_1) | 10 µM | 2.5 | 0.5 µM |
| Primer (r_mdnA_1) | 10 µM | 2.5 | 0.5 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 8.0 | 0.7 ng |
| Polymerase | 1 | 5 U |
in addition MnCl2 and H2O as shown following
| Sample number | MnCl2 in µl | Final concentration MnCl2 | H2O in µl |
|---|---|---|---|
| 1 | 0 | 0 | 23.5 |
| 2 | 0.5 | 0.05 mM | 23.0 |
| 3 | 1.0 | 0.1 mM | 22.5 |
2. Temperature profile
- see 12th labday
- Eppendorf Mastercycler personal, program IGMDNA1
Results:
- 3 PCR products containing random mutated mdnA
Further tasks:
- Verification of these PCR products by agarose gel electrophoresis
- Extraction and purification these (positive) samples
- Enzyme digestion using NarI and AatII
- Heating the half of each samples 10-20 min by 65°C and the other half not
- Verification using agrose gel electrophoresis
Midiprep of pJC354_WZA2 (TorA-bla vector) carrying cells
Investigators: Sebastian, Paul
Aim:
- Midiprep of pJC354_WZA2 carrying cells using Quiagen Plasmid Plus Midi Kit
Materials:
- Qiagen Plasmid Plus Midi Kit
- over night cultures ###
Output: DNA Eppi Tube label (-20°C), Concentration: ~1mg/ml
Further tasks:
- Annealing of oligos for cleavage sites
- Digest of vector with XhoI and NheI
- Ligation of annealed oligos into the vector
25th Labday 2011-06-28
Agarose gel electrophoresis of mutated mdnA
Investigators: Katharina
Aim: Verification of random mutated mdnA
Time: 2011-06-26,12:45-13:30
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- 3 PCR products of mutated mdnA
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 30 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler
3. Run
- 100 V
- time: 1 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | PCR sample 1 | 50 | |
| 3 | PCR sample 2 | 50 | |
| 4 | PCR sample 3 | 50 |
Gel extraction of mutated mdnA
Investigators: Katharina
Aim: Extraction of mdnA, which was amplified and mutated (using error prone PCR)
Time: 2011-06-26,13:00-14:00
Materials:
- NucleoSpin Extract II (Macherey-Nagel) extraction Kit
- PCR products 1 and 2 containing mutated mdnA
Method:
- performing gel extraction based on manufacturer's protocol
- elute in 30 µl buffer NE
Results:
- purified PCR products of mutated mdnA (1 and 2)
Further tasks:
- restriction enzyme digestion with purified mdnA using AatII/ NarI
- Verification by agarose gel electrophoresis
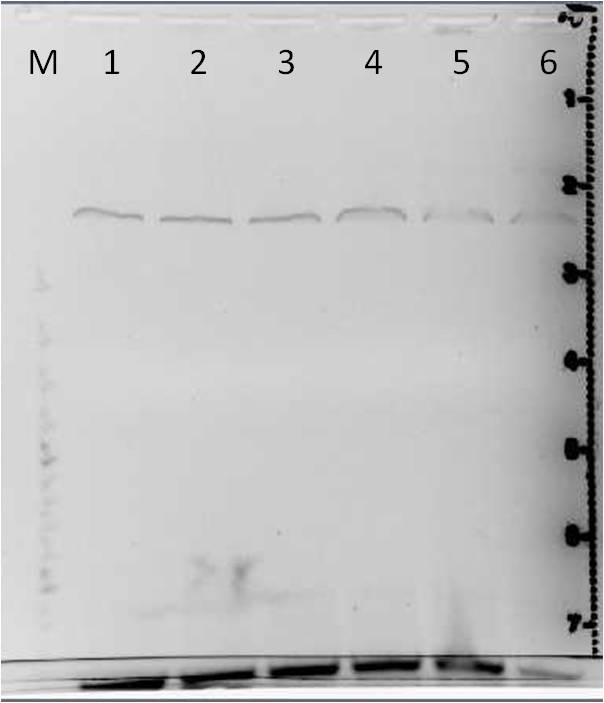
Restriction enzyme digestion and agarose gel electrophoresis of mutated mdnA
Investigators: Katharina
Time: 2011-06-28,14:00-17:00
Aim: Restriction enzyme digestion of random mutated mdnA and verification using gel electrophoresis
Materials:
- Restriction enzymes: AatII, NarI
- NEB Buffer 4
- PCR products of random mutated mdnA(1 and 2), purified from gel
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Gene Ruler, 100bp plus, DNA Ladder (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Restriction enzyme digestion
- 5 µl of purified PCR product were left undigested
| components | volume of PCR products /µl| |
| DNA | 25 |
| NEB Buffer 4(10x) | 4 |
| Enzyme AatII | 1 |
| Enzyme NarI | 1 |
| H2O | 9 |
| Total volume | 40 |
- digestion for 1 h
- after digestion 17.5 µl of digested PCR product were taken for heat inactivation of the restriction enzyme (60°C for 20 min)
2. Production of 1 % agarose gels
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
3. Loading samples
a. restriction enzyme digestion products of random mutated mdnA
- 35 µl PCR product and 5 µl 6x loading dye
- 2 µl DNA ladder gene ruler, 100bp plus (Fermentas)
4. Run
- 120 V
- time: 45 min
Results:
File:UP Digestion Error Prone 2010-28-06.png
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | marker | 2 | |
| 2 | undigested mutated mdnA sample 1 | 5 | |
| 3 | digested mutated mdnA sample 1 | 17.5 | |
| 4 | digested and heat inactivated enzyme after digestion of mutaded mdnA sample 1 | 17.5 | |
| 5 | undigested mutated mdnA sample 2 | 5 | |
| 6 | digested mutated mdnA sample 2 | 17.5 | |
| 7 | digested and heat inactivated enzyme after digestion of mutaded mdnA sample 2 | 17.5 |
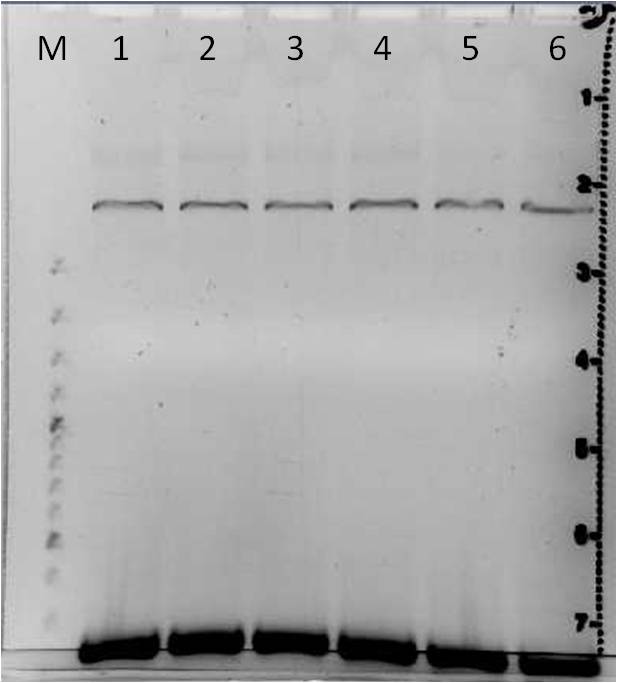
Error prone PCR (varied MnCl2 concentration) of mdnA
Investigators: Steffi
Time: 2011-06-28,15:00-15:50
Aim: Random mutation of mdnA gene based on increased MnCl2 concentration
Materials:
- Genaxxon Bioscience Taq Polymerase
- 10x Genaxxon Bioscience Taq Polymerase buffer containing 15 mM MgCl2
- dNTPs (10 mM)
- forward primer: sf_mdnA_1 (10 µM)
- reverse primer: r_mdnA_1 (10 µM)
- template DNA pARW089 (857,3 ng/ µl measured by Nanodrop a second time)
- Genaxxon Bioscience MgCl2 stock solution (50 mM)
- MnCl2
- Eppendorf Mastercycler personal, program IGMDNA1
Error prone PCR of mdnA
- increasing MnCl2 concentration
1. Mix for error prone PCR with increasing MnCl2 concentration
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| Genaxxon Bioscience Taq Polymerase buffer containing MgCl2) | 15 mM (MgCl2) | 5 | 1.5 mM MgCL2 |
| MgCl2 | 50 mM | 5.5 | 5.5 mM |
| dNTPs | 10 mM | 2.0 | 0.4 mM |
| Primer (each) | 10 µM | 2.5 | 0.5 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 5.58 | 0.7 ng |
| Polymerase | 1 | 5 U |
2. Complete pipet scheme
| Component in µl | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Template DNA | 8 | 8 | 8 | 8 | 8 | 8 |
| dNTPs | 2 | 2 | 2 | 2 | 2 | 2 |
| 10x Genaxxon polymerase buffer (with MgCl2) | 5 | 5 | 5 | 5 | 5 | 5 |
| MgCl2 (50 mM) | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 |
| MnCl2 (5 mM) | 0 | 0.5 | 1 | 1.25 | 1.5 | 2 |
| Forward primer (sf_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Reverse primer (r_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| H2O | 23.5 | 23.0 | 22.5 | 22.25 | 22.0 | 21.5 |
3. Temperature profile
- see 12th labday
- Eppendorf Mastercycler personal, program IGMDNA1
Results:
- 6 PCR products containing mutated mdnA
Further tasks:
- Verification of these PCR products by agarose gel electrophoresis
- Purification and cloning of positive samples
- Sequencing
26th Labday 2011-06-29
5l LB Media (liquid)
Investigators: Sebastian, Niels
Aim: Generating media for further research
Materials:
- Yeast (5 g/l)
- Tryptone (10 g/l)
- NaCl (10 g/l)
- autoclaved
oligo hybridization for TEV and 14_3C protease cleavage sites
Investigators: Sebastian, Paul
Materials:
2 reaction batches: of_NheI_CS-TEV_XhoI (Cleavage site for TEV) and of_NheI_143C-cleavage_XhoI (Cleavage site for 14_3C)
- 1 µl Oligo 1(of_NheI_CS-TEV_XhoI or of_NheI_143C-cleavage_XhoI forward)
- 1 µl Oligo 2(of_NheI_CS-TEV_XhoI or of_NheI_143C-cleavage_XhoI reverse)
- 4 µl 100mM TrisHCl pH 8
- 8 µl 5 mM MgCl2
- 26 µl H2O
- Eppendorf Mastercycler personal, program ORIGAMI:
1 96°C 7 min
2 96°C 1 min
-1°C R=0,3°/s
goto 2 rep 70
hold 4°C
Restriction of pJC354_WZA2-vector
Investigators: Sebastian, Paul, Sascha
Materials:
- XhoI (NEB), NheI (NEB)
- 100x BSA (NEB), 10x buffer 4 (NEB)
- pJC354_WZA2-vector
Protocol:
- 1µl XhoI
- 1µl NheI
- 5µl 10x buffer 4
- 0,5 µl vector [~500ng]
- 0,5 µl 100x BSA
- 42 µl pure H2O
- Reaction for 2h at 37°C
Further tasks:
- preparative agarose electrophoresis
- Ligation of digested vector with annealed oligos (over night, 18°C)
preparative agarose electrophoresis
Investigators: Sebastian, Paul, Sascha
Method:
Samples digested with NheI and XhoI
vector: digested pJC354_WZA2 source: Janina; prep-type: midi-prep ; prep-date:24.06.11 ; clone-no: 6
- control undigested pJC354_WZA2 source: Janina; prep-type: midi-prep ; prep-date:24.06.11 ; clone-no: 6
Electrophoresis
sample preparation: 50 µl pJC354_WZA2 (digested) + 10 µl 6x loading dye solution, 1 µl pJC354_WZA2 (undigested) + 1 µl 6x loading dye + 4 µl dest. water
0,5 g Agarose, 50 ml TAE (1%), 2 µl GELRED, at 115 Volt,running time:_?___ minutes
marker: GeneRuler ladder mix (Fermentas)
| lane | Sample | Sample/µl | Expected size (bp) | approx. size |
|---|---|---|---|---|
| 1 |
marker |
6 µl |
||
| 2 |
pJC354_WZA2 (digested) |
~40 µl | 130, 4700 | 130, 8000, 2400 |
| 3 |
pJC354_WZA2 (digested) |
~20 µl | 130, 4700 | 130, 8000, 2400 |
| 5 |
pJC354_WZA2 (undigested) |
6 µl | 4000 for supercoiled, 4700 for not supercoiled plasmid |
2400,2400-3000, 7500 |
Gel Extraction
DNA excision: lane 2 and 3: 8000 bp band;
DNA purification: with QIAquick Gel Extraction Kit according to Qiagen manual (incl. isopropanol step)(done by Tobias)
Result: size estimation by eye;
lanes 2+3: plasmid was digested, small band has the expected size, two upper bands with unexpected size;
lanes 5: plasmid was undigested; 7500 bp unexpected; 3000 bp band might be undigested supercoiled plasmid
Conclusions:
- exact vector fragment unknown, therefore 8000 bp band was
- because of unexpected sizes marker should be compared with
another marker, plasmids should be digested to yield several fragments
for size control, maybe heat inactivation might help to resolve higher bands
Output: one plasmid fragments with 8000 bp in 1,5 ml eppis,
stored at -20 °C, box with fragments
PCR for amplification of mdnA with C-terminal myc-tag
Investigators: Jessica
Aim: amplificate mdnA of pARW089 and add C-terminal myc-tag with primers pf_mdnA_mycC_XbaI and pr_mdnA_mycC_PstI_NotI_SpeI
Time: 2011-06-29, 14:00-18:00
Materials:
- NEB OneTaq DNA Polymerase
- 5x Buffer NEB OneTaq
- dNTPs (10 mM)
- forward primer: pf_mdnA_mycC_XbaI (10 µM)
- reverse primer: pr_mdnA_mycC_PstI_NotI_SpeI (10 µM)
- template DNA pARW089
- Eppendorf Mastercycler gradient, program SABRINA
50 µl reaction
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| 5x Buffer NEB OneTaq | 5x | 10 | 1x |
| dNTPs | 10 mM | 1 | 0.2 mM |
| Primer (each) | 10 µM | 1 | 0.2 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 5 | ~0.7 ng |
| NEB OneTaq DNA Polymerase | 0.25 | 1.5 U | |
| H2O | 31.75 |
Temperature profil
| Temperature in °C | Time in s | |
|---|---|---|
| 94 | Hold | |
| 94 | 60 | Initial denaturation |
| 94 | 20 | Denaturation |
| 60 | 20 | Annealing |
| 72 | 20 | Elongation |
| --> 15 cycles | ||
| 94 | 60 | Denaturation |
| 70 | 40 | Annealing |
| 72 | 20 | Elongation |
| --> 15 cycles | ||
| 70 | 300 | Final extension |
| 6 | Hold |
Further tasks:
do gel electrophoresis
PCR of mdnA for phage display (strategy 1) repeated with new ordered forward primer
Investigators: Sandrina
Time: 2011-06-29,14:00-18:00
Aim: amplification of mdnA with SfiI restriction sites (vector pARW089) to clone it into pAk100 bla KDIR
Materials/Methods:
see 2011-06-10
changes:
- elongation time 40 s
- template 5 µl
- program: Sabrina, eppendorf master cycler gradient
further tasks:
gel electrophoresis with PCR product
Error prone PCR (varied MnCl2 concentration) of mdnA
Investigators: Katharina
Time: 2011-06-29,12:00-13:00
Aim: Random mutation of mdnA gene based on increased MnCl2 concentration
Materials:
- Genaxxon Bioscience Taq Polymerase
- 10x Genaxxon Bioscience Taq Polymerase buffer containing 15 mM MgCl2
- dNTPs (10 mM)
- forward primer: sf_mdnA_1 (10 µM)
- reverse primer: r_mdnA_1 (10 µM)
- template DNA pARW089 (857,3 ng/ µl measured by Nanodrop a second time)
- Genaxxon Bioscience MgCl2 stock solution (50 mM)
- MnCl2
- Eppendorf Mastercycler personal, program IGMDNA1
Error prone PCR of mdnA
- increasing MnCl2 concentration
1. Mix for error prone PCR with increasing MnCl2 concentration
| Component | Stock concentration | Volume in µl | Final concentration |
|---|---|---|---|
| Genaxxon Bioscience Taq Polymerase buffer containing MgCl2) | 15 mM (MgCl2) | 5 | 1.5 mM MgCL2 |
| MgCl2 | 50 mM | 5.5 | 5.5 mM |
| dNTPs | 10 mM | 2.0 | 0.4 mM |
| Primer (each) | 10 µM | 2.5 | 0.5 µM |
| DNA (diluted 1:100) | 8.57 ng/ µl | 5.58 | 0.7 ng |
| Polymerase | 1 | 5 U |
2. Complete pipet scheme
| Component in µl | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Template DNA | 8 | 8 | 8 | 8 | 8 | 8 |
| dNTPs | 2 | 2 | 2 | 2 | 2 | 2 |
| 10x Genaxxon polymerase buffer (with MgCl2) | 5 | 5 | 5 | 5 | 5 | 5 |
| MgCl2 (50 mM) | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 |
| MnCl2 (5 mM) | 0 | 0.5 | 1 | 1.25 | 1.5 | 2 |
| Forward primer (sf_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| Reverse primer (r_mdnA_1) | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| H2O | 23.5 | 23.0 | 22.5 | 22.25 | 22.0 | 21.5 |
3. Temperature profile
- see 12th labday
- Eppendorf Mastercycler personal, program IGMDNA1
Results:
- 12 PCR products (each sample was prepared twice) containing mutated mdnA
Further tasks:
- Verification of these PCR products by agarose gel electrophoresis
- Purification, digestion and cloning of positive samples
- Sequencing
27th Labday 2011-06-30
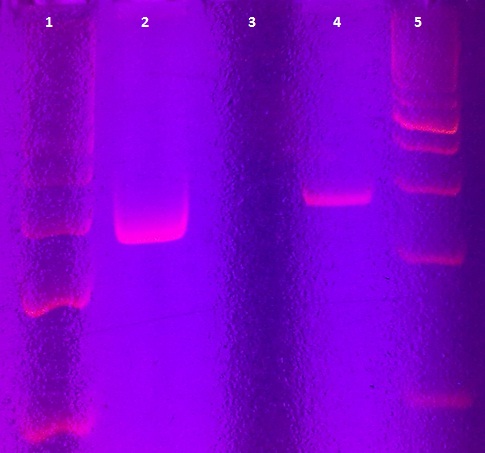
Agarose gel electrophoresis of myc-tagged mdnA and mdnA for phage display
Investigators: Nadja, Jessica, Sandrina
Aim: Verification of myc-tagged mdnA and mdnA for phage display (strategy 1)
Time: 2011-06-30,10:00-12:00
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- PCR products: myc-tagged mdnA, mdnA for phage display (strategy 1) from 2011-06-29
- DNA Ladder Gene Ruler, 100bp plus (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 5 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler (1:10)
3. Run
- 115 V
- time: 1 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker | 2 | |
| 1 | myc-tagged mdnA | 6 | 233 |
| 2 | mdnA with SfiI | 6 |
Agarose gel electrophoresis of error prone PCR of mdnA from 2011-06-29 and gel extraction
Investigators: Nadja, Jessica
Aim: Verification of random mutated mdnA
Time: 2011-06-30, 12:00-17:00
Materials:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- 12 PCR products: sample 1a to 6b from 2011-06-29
- DNA Ladder Gene Ruler, 100bp plus (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- mAgarose = 0.5 g in 50 ml 1x TAE
- adding 2 µl gel red
2. Loading samples
- 30 µl PCR product and 6x loading dye
- 2 µl DNA ladder gene ruler (1:10)
3. Run
- 115 V
- time: 1 h
Results:
| gel 1 | gel 2 | ||||||
|---|---|---|---|---|---|---|---|
| lane | Sample | Volume in µl | Expected size in bp | Sample | Volume in µl | Expected size in bp | |
| M | marker | 2 | marker | 2 | |||
| 1 | PCR sample 1a | 36 | 276 | PCR sample 4a | 36 | 276 | |
| 2 | PCR sample 1b | 36 | 276 | PCR sample 4b | 36 | 276 | |
| 3 | PCR sample 2a | 36 | 276 | PCR sample 5a | 36 | 276 | |
| 4 | PCR sample 2b | 36 | 276 | PCR sample 5b | 36 | 276 | |
| 5 | PCR sample 3a | 36 | 276 | PCR sample 6a | 36 | 276 | |
| 6 | PCR sample 3b | 36 | 276 | PCR sample 6b | 36 | 276 | |
Gel Extraction
- DNA excision: PCR products 1a to 6b
- DNA purification: with Qiagen Gel Extraction Kit according to Qiagen manual (incl. isopropanol step, elute in 30 µl H2O)
Results:
- DNA concentrations measured with Nanodrop were lower than 10 ng/µl
Further tasks:
- repeat with remaining PCR samples?
Repeated PCR of mdnA for phage display (strategy 1) repeated with new ordered forward primer
Investigators: Sandrina, Sabine
Time: 2011-06-30,15:00-18:00
Aim: amplification of mdnA with SfiI restriction sites (vector pARW089) to clone it into pAk100 bla KDIR
Materials/Methods:
see 2011-06-10
changes:
- elongation time 40 s
- template 5 µl
- program: Sabrina, eppendorf master cycler gradient
further tasks:
gel electrophoresis with PCR product
 "
"