Team:Harvard/Template:NotebookDataJuly
From 2011.igem.org
Contents |
July 1st
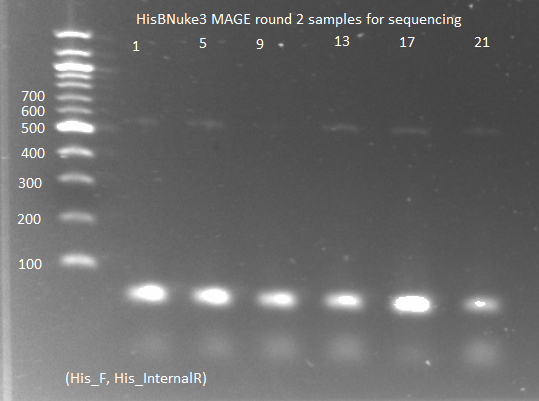
HisBNuke3 MAGE results:
- All the MAGE round 1 colonies grew overnight in the LB but not the NM medium: either they all had the HisB gene knocked out (unlikely) or there is something wrong with the NM medium. As a control we grew (∆HisB)∆pyrF∆rpoZ+pKD46 in NM, but nothing grew, implying that the problem is with the NM medium.
- There were small colonies on the MAGE round 2 amp plates, so we chose 12 from each plate and grew them in 100µL of SOC medium.
- PCR of the colonies for sequencing:
- KAPA mastermix and protocols, with His3_F and His3_internalR primers, 2µL culture as template, 65˚C annealing, 15 sec elongation, 25 cycles
- ran on gel: band around 500-600, but very faint (probably because we did not let the culture grow up enough)
- sent to Genewiz to be sequenced with His3_F primer
Lambda Red recombination results:
- The plates from 6/30 had an almost crystalline formation on them, which may or may not be bacteria. The 6/29 plates, which we had left at 30˚C, did have colonies but they may be contaminants. We grew up 12 colonies from each 6/29 plate and 6 "colonies" from the 2mL 6/30 plate in SOC medium with kanamycin, and next week we will PCR the 1529620 locus to check.
pZE21G spec backbone:
- PCR of backbone was successful (see E gel below). Gel extracted with Qiagen kit with the following modifications to the kit instructions:
- 2 gel volumes of buffer QG, heated 10-15 min
- after heating added 10µL 3M NaOAc
- eluted in 30µL ddH2O
- 157.4ng/µL 260/280=1.9; 129.2ng/µL 260/280=1.88
Selection system plates:
- Made 3-AT and 5-FOA plates following 2006 Meng Nature Protocol (see protocols for details). 5-FOA plates have a blue stripe, 3-AT plates (0.5 mL 3-AT) have a green stripe.
July 4th
FIREWORKSJuly 5th
Team ZF Assembly
- Assemble and confirm zif268 plasmid
- Lac-ω-zif268 with spec → Lac-zif-ω with cpic
- ZF: 2-3 ultramers
- PCR out ω subunit
Team Wolfe Selection
Goals for the week:
- MAGE out HisB
- What we've been trying/why it's not working — keep trying!
- PCR out Kan Cassette (colony PCR)- analysis/check that it worked
- We did the insertion and have things growing in Kan that look like our cells — we probably have it!
- As long as it's the right size and in the right place, it probably worked, meaning we have 2 HIS genes → MAGE is to remove the endogenous genome copy in vivo
- Construct (PSR01): ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~
- Fix/Test NM media
- Made last week, nothing worked in it — try more glucose
- Wolfe strain seems to grow really slowly (~36h); try glycerol stock of PKD46, grows faster
- Make other 3-AT plates (1.5)
- 3-AT is a competitive inhibitor of His; the stronger His is expressed, the more 3-AT the cell can tolerate and still grow
- Test 3-AT plates
Today's results:
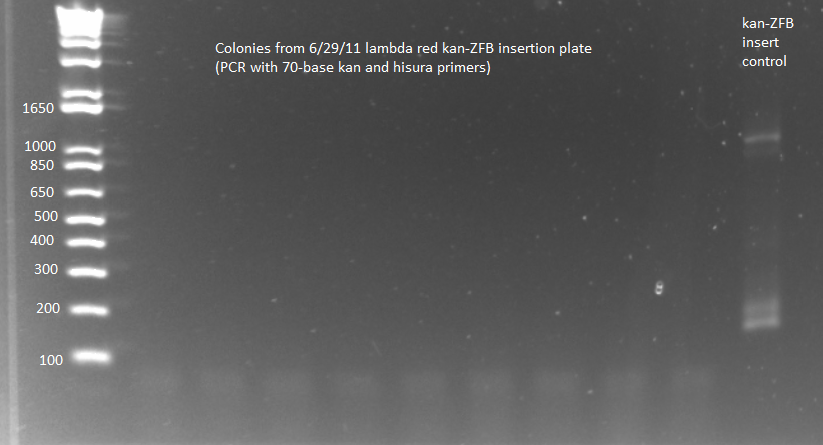
- PCR to check whether kan-ZFB-wp-hisura was successfully inserted into the genome
- Used KAPA master mix and protocols, same procedure as 6/29/11
- Because of KAPA shortage, only ran samples 6-8 from 100µL 6/29 plate and 6-7 from 2mL 6/29 plate plus (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture as a control (should produce a band around 300bp, while the insert would make the band 2-3kb)
- Ran on E-Gel with team TolC's samples, but there was an issue loading the gel, so also ran separately on another gel. Unfortunately most of the PCR reactions strangely did not work, but two of the samples appeared not to have the insert (see below).
- Redo PCR:
- out of KAPA so had to use Invitrogen Platinum Supermix
- each reaction had 22.5µL supermix, 0.5 µL each 10µM primer, 1µL template, 0.5µL H2O
- 20 reactions, including one from the same control used above.
- followed Invitrogen instructions, with 3 min elongation, 56 C annealing, 30 cycles
- Assuming the PCR is unsuccessful, we will grow up (ΔHisB)ΔpyrFΔrpoZ+pKD46 (from 6/28 plate) overnight in LB+amp
- NM medium:
- To test Sarah's NM (made 6/30) we will grow up pKD46 strain in 3mL of it overnight at 30 C. If no bacteria grew, we will try remaking it ourselves.
Team TolC
- PCR out and insert Kan construct in front of TolC (similar to Wolfe selection)
- Construct 1: ~ - KAN - ZFbs - weak promoter - ~ (done, PCR, gel extraction)
- Used primers ZFB-wp-tolC-r and tolC-kan-f
- Construct 2: ~ TolC ~ (do not have primers yet)
- Note that ~ indicates a homology region
- Construct 1: ~ - KAN - ZFbs - weak promoter - ~ (done, PCR, gel extraction)
- Need the EcNR2 strain to come in before we can work with it (came in Thursday 7/7/2011, the one from Wednesday 7/6/2011 was not growing well.)
- Inactivate rpoZ (similar to inactivating His3, but should be easier to use MAGE with- EcNR2 is a strain designed for MAGE)
- MAGE oligo rpoZ deletion will arrive on Thursday 7/7/2011
- Make SDS (how we test TolC +vely)(done, stored at room temperature)
- Only cells with active TolC can survive SDS treatment
Web Design
To Do
- Safety Description
- Email Alain/Harvard biosafety group ✔ (Meeting pending)
- Needed information:
- Strain names
- Antibiotic Resistance
- DNA inserts/removals
- Draft Project Description ✔
- Finalize
- Update the Facebook page/create a new 2011 page
- Get the Public Wiki up and running
- Figure out templates
- Update Twitter
Notes to All Teams
- Teams: Make sure everything is organized in your dropbox!
- *Announcing a New (and Final) Plasmid Name* PSR01: formerly known as Wolfe Plasmid/Vatsan's Plasmid/Selection
- Contains ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~
July 6th
Team ZF Assembly
Because our primers still haven't arrived, the members of Team ZF Assembly are working with the Web Design Team.
Team Wolfe Selection
- Redo PCR Results:
- Samples 1,3,4,5,6,7,8,9,11 from the 100µL 6/29 plate all showed unsuccessful introduction of kan-ZFB-wp-hisura
- Samples 2,3,4,6,7,8,9,10,11,12 from the 2mL 6/29 plate all showed unsuccessful introduction of kan-ZFB-wp-hisura
- (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture used as a control
- NM media has 1% of the glucose concentration written in the protocol, it is 0.4mg/ml instead of 40mg/ml. Nevertheless, our overnight culture of pKD46 grew successfully so we will continue to use the media unless other problems arise.
- One possible explanation for our failure to PCR out a kan-ZFB insert from the recombined colonies is that the insert was integrated into the genome somewhere else besides the 1529620 locus. This might be why the PCR shows no insert while the colony still grows in kanamycin. We will try to PCR the insert using primers that anneal to the kan and his-ura sections (since these primers are about 70 bases long, we will do a 2-step cycle)
- Invitrogen Platinum Supermix reagents and protocols, with 3:30 at 72˚C instead of separate annealing and elongation, 25 cycles
- used same cultures as for PCR listed above
- kan-ZFB-wp-hisura insert as control
- results from 2 E Gels: PCR not very successful. Colonies showed no bands while the control showed only a side product too small to be the full insert.
- Grew up 24 more kan-ZFB lambda red recombined colonies (12 from 100µL 6/29 plate, 12 from 2mL 6/29 plate) in LB+kan in 96 well plate for PCR tomorrow
- Diluted 24 colonies (1 ss+pKD46 control, 11 from MAGE round 2 1µL plate, 12 from MAGE round 2 10µL plate) in 30µL ddH2O and split between NM,NN+Histidine, and LB+amp to check his phenotype
- NM+his media: 49mL NM, 1 mL 5% histidine solution
- Made 1.5mL 3-AT plates
Team TolC
- Gel extraction of KAN-ZFBs-wp as there is a side product.
- To increase yield, spin down the contents of both eppendorf tubes into a single column, have a 2:1 volume ratio of QG Buffer to gel volume, e.g. 1ml QG buffer to 0.5ml gel in step 2, and elute in 30µl water in step 13)
- checked the ECNR2 strains, both appear to have not grown
- note from 7/7/2011, new ECNR2
- running another PCR since there was errors in gel extraction, and the yield was less than 20ng/µl, and purity was very bad.
TOLCINV PCR-Machine 7
- 94°C for 2 min
- 94°C for 30 s
- 53°C for 30 s
- 72°C for 1.5 min
- Previous three steps repeat 30 times
- 72°C for 5 min
- 4°C forever
Web Design
- Project description
- Web design brainstorming;exploring past designs and templates
Retrieved from "http://2011.igem.org/Team:Harvard/Template:NotebookDataJuly"
 "
"