Team:Freiburg/Notebook/19 July
From 2011.igem.org

Contents |
green light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
blue light receptor
Theoretical Gibson-Assembly
Investigators: Sandra, Sophie
Primer Degin for Blue light sensor (lovTAP) + trp promotor (BBa_K322999) and tetR Gen + tetO promotor (BBa_Q04400).
- LOVtap-Trp_up: gaattcgcggccgcttctagtcacacaggaaagtactatgt
- LOVtap-Trp_dw: tacttttatctaatctggacatctagtatttctcctctttgtcgataccctttttacgtg
- TetR-TetO_up: aaagaggagaaatactagatgtccagattag
- TetR-TetO_dw^: ctgcagcggccgctactag
^we also have another primer for this one, but we are not sure about the length/melting temperature. Might be changed.
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
PCR
| Name: Ruediger
| Date: 19.07.11 |
| Continue from Experiment (Date)
PCR 18.07 (Name) | |
| Project Name: GFP Pbd | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | P18, P19, P20 |
| 2.5µl | Primer dw | P28 |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | PCR product of P1,P3,S14 from yesterday |
| 0.5 µl | Phusion (add in the end) |
What program do you use?
One set of probes was prepared and then split into 3 tubes each to test them at Annealingtemperatures 44C, 52C and 60C for the first 10 cycles. Then Annealingtemperature 60C for 25 cycles
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
S14+P18+P28
S14+P19+P28
S14+P20+P28
Stored in PCR product box
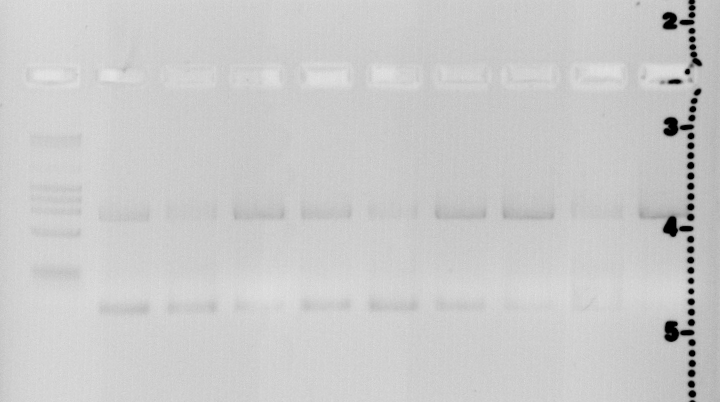
Lane1 Quick Load Marker Lane2 44C S14+P18+P28 Lane3 44C S14+P19+P28 Lane4 44C S14+P20+P28 Lane5 52C S14+P18+P28 Lane6 52C S14+P19+P28 Lane7 52C S14+P20+P28 Lane8 60 S14+P18+P28 Lane9 60 S14+P19+P28 Lane10 60 S14+P20+P28
Digestion
| Name: Ruediger
| Date: 19.07 |
| Continue from Experiment (Date) 19.07 PCR
(Name) Ruediger | |
| Project Name:
GFP Pbd | |
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Vector (ratio 1:3 to insert)
Inserts (500ng)
| Components | Vector (μl) | Insert1 (μl) | |||
| DNA (500ng) | 5,6 | 12,5 | 4,7 | 3,5 | 3,8 |
| BSA (100x) (5μl) | |||||
| NEB4 Buffer (5μl) | |||||
| Enzyme 1 (1μl) | SpeI | SpeI | XbaI | XbaI | XbaI |
| Enzyme 2 (1μl) | PstI | PstI | PstI | PstI | PstI |
| H2O (38 μl- DNA) | 32,5 | 25,5 | 33,7 | 34,5 | 34,2 |
| In total 50 μl | |||||
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample | DNA concentration (μg/μl) |
| S14+P20+P28 (short:20) | 132 |
| S14+P19+P28 (short 19) | 145 |
| S14+P18+P28 (short 18) | 107 |
| S39 | 90 |
| S43 | 40 |
Documentation:
Why are you doing this experiment? Where are the samples stored? Name of samples, antibiotica resistance, vector used etc.
| Want to ligate GFPpbd (3 different versions P18/19/20) into PR vectors (one with strong Promotor strong RBS, one with middle Promotor, middle RBS)
CM Resistance Mistake: took 100x BSA instead of 10X |
 "
"
 Contact
Contact