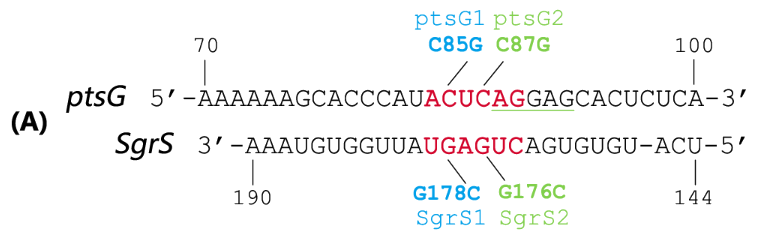Team:Peking S/project/nonb
From 2011.igem.org
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Non-Boolean Dynamics
Synthesizing a Modular Comparator Using Small Regulatory RNAs
Introduction
Our ‘chemical wire’ toolbox is not only applicable to Boolean logic gene networks, but also amenable to non-Boolean population dynamics, for instance, the microbial population density balancer mentioned above.
In such a dynamical network, we need to construct a comparator device that can integrate two environmental signals. We managed to fulfill the task with the simplest elements and types of interactions of gene regulation, i.e., the inducible promoters, mRNAs and regulatory small RNAs that silence them. We will demonstrate that simply by combining these three types of genetic components, a comparator module can be implemented and thus to work in population balancer system to verify the feasibility of our "chemical wire" toolkit for non-Boolean population dynamic.
Design
Small noncoding RNAs (sRNAs) have been depicted in recent years to play central roles in gene regulation. The special features of small RNA mediated regulation are fast responsive, noise insensitive and threshold effect, which inspire us to construct an artificial comparator based on small RNA regulation. Before biological implementation in bench, during circuit design, series of requirements must be met.
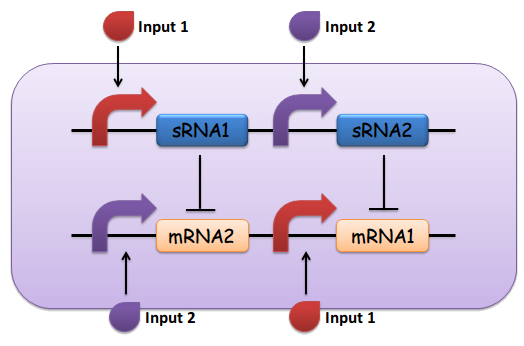
(1) Input 1 activates mRNA and sRNA 1.
(2) Input 2 activates mRNA and sRNA 2.
(3) Suppose there is more input 1, sRNA 1 will strongly repress mRNA 2 and vice versa.
(4) Suppose there is less input 2, the repression going towards the other way should be weak enough. This will lead to the production of output 1, and little to no production of output 2.
(5) The output of the device is either dominated by output 1 or output 2. By seeing which is the case, we can infer whether input 1 or input 2 was present in greater amounts. (See Fig. 1)
Fig.1 The design of the sRNA-dependent comparator, as described above
Numerous sRNA/5’UTR pair have been reported in literature. We regarded the ptsG/SgrS pair as an ideal candidate for our comparator design for its special regulation mechanism described below.
Mechanism
SgrS is an Hfq-binding small antisense RNA that is induced upon phosphosugar stress (Vanderpool, 2007). It forms a ribonucleoprotein complex with RNase E through Hfq to mediate silencing of the target ptsG mRNA encoding the major glucose transporter (Geissmann and Touati, 2004). A 31-nt-long stretch in the 3’ region of SgrS is partially complementary to the translation initiation region of ptsG mRNA, and a 6 nt region overlapping the Shine-Dalgarno sequence of the target mRNA turns out to be crucial for SgrS’ function, shown as Fig 2 (Kawamoto et al., 2006; Maki et al., 2010).
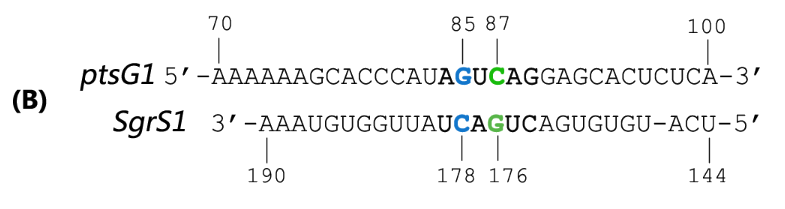
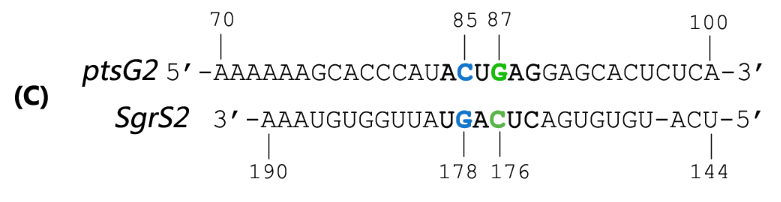
Fig. 2 Sequence alignment of wild type ptsG/SgrS pair and its mutant complementary pairs.
(A) The partial complementary region of ptsG (wt) mRNA and its corresponding sRNA SgrS.
(B) The complementary pair of ptsG1 mRNA and corresponding SgrS1.
(C) Another complementary pair site-mutant version of ptsG2 mRNA and corresponding SgrS2.
Teppei Morita et.al’ s work suggests that two mutations (C85G and C87G) in ptsG mRNA could completely impair the ability of SgrS to downregulate its expression, while compensatory mutations of SgrS (G178C and G176C) restore the gene silencing ability. These results indicate that it is the base pairing of the two RNAs rather than particular nucleotides that is important for SgrS action. They have also illustrated that sequence outside this region, even though complementary, is rather dispensable for the efficient silencing (Kawamoto et al., 2006). This makes mutant ptsG/SgrS pairs orthogonal to genetic context of the host cell.
 "
"