Team:Freiburg/SampleData
From 2011.igem.org

Contents |
Sample Data
How Our System Works
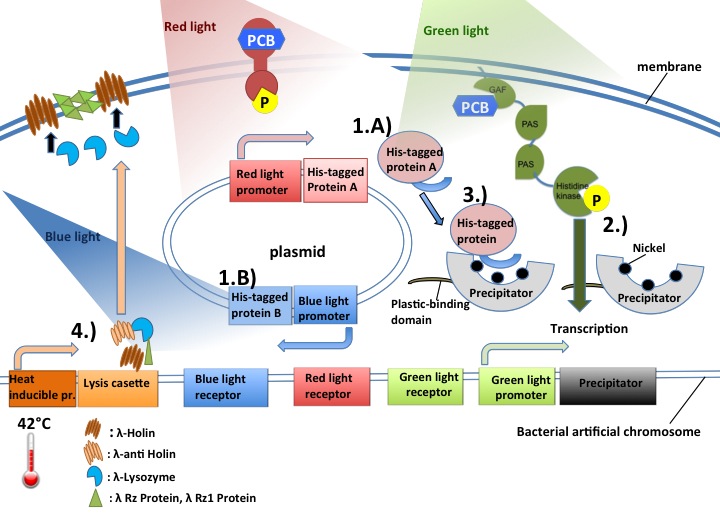
How our system works:
- 1.A) Exposure of bacterial culture to red light (705 nm)
- Red light inducible promoter leads to expression of His-tagged protein of interest.
- 1.B)Exposure of bacterial culture to blue light (470nm)
- Blue light leads to a conformational change of the blue light receptor (LovTAP) and the expression of a His-tagged protein.
- 2.) Exposure of bacterial culture to green light (532 nm)
- Green light inducible promoter leads to expression of the "Precipitator".
- 3.) Precipitator-Protein of interest complex
- His-tag of the protein of interest is bound by the Nickel "Precipitator" complex.
- 4.) Cell lysis
- Cells are autolysed by simply being heated for about 1 hour to 42°C.
And thus the Precipitator-Protein of interest complex is in solution. It can now bind to the surface of serological pipettes (made of polystyrene) with the help of a hydrophobic plastic binding domain, and stabilize the protein of interest long enough to endure two wash steps to elute it.
Our Favorite New Parts
1.
[http://partsregistry.org/Part:BBa_K608408 BBa_K608408] GST
The GST-tag was PCR amplified from a pGEX vector with overhang primers including the iGEM restriction sites and then pasted into the iGEM vector. To verify the functionality of the construct we cloned it before a GFP sequence and expressed it with an IPTG inducible vector. Results see partsregistry page. The submitted sequence was partially confirmed by sequencing.
2. [http://partsregistry.org/Part:BBa_K608406 BBa_K608406]
Precipitator
The Precipitator is a new artificially designed LRR protein. It is meant as protein that binds Nickel ions with Histidines grouped on its surface. The bound Nickel can then precipitate His-tagged proteins. In our Lab in a Cell it should function as an adaptor between the plastic surface of pipettes and the His-tagged protein. Please look at our detailed description of the design layout in our modeling section. The sequence was synthesized and cloned into the iGEM vector. The submitted sequence was fully confirmed by sequencing.
3. [http://partsregistry.org/Part:BBa_K608404 BBa_K608404] IPTG-inducible Promoter with plastic binding domain-tagged GFP
Also new parts
[http://partsregistry.org/Part:BBa_K608101 BBa_K608101] CcaR, green light response regulator
[http://partsregistry.org/Part:BBa_K608102 BBa_K608102] CcaS, green light receptor
Pre-existing Parts
Lysis cassette
The Concept
The idea behind our lysis cassette was to be able to achieve cell lysis by simply heating the bacteria to 42°C for a short period of time. This was to be accomplished by combining the biobricks [http://partsregistry.org/Part:BBa_K098995 BBa_K098995] (temperature sensitive promoter) and [http://partsregistry.org/Part:BBa_K124017 BBa_K124017] (Bacteriophage λ lysis genes). Sequencing of these parts (carried out by GATC Biotech GmbH) showed some fundamental inconsistencies leading to a lot of time spent on fixing them.
The sequences were corrected and the new biobricks [http://partsregistry.org/Part:BBa_K608351 BBa_K608351] and [http://partsregistry.org/Part:BBa_K608352 BBa_K608352] were sent to the registry, although the novel temperature sensitive lysis cassette composite's sequence was verified some 18 hours before the Wiki-freeze, so that it could not be sent to the registry on time. Nevertheless, some preliminary OD-measurement results did hint at the expected functionality of the newly developed composite part.
For more Information, please check our description page https://2011.igem.org/Team:Freiburg/Description#Part_Design_2
[http://partsregistry.org/Part:BBa_K608151 BBa_K608151] translational unit of pcyA
We designed this composite in order to gain the enzyme pcyA (phycocyanobilin-ferredoxin oxidoreductase),
this design is necessary if the part is going be integrated into an assembly of various genes.
The enzyme plays an important role in the production of the PCB chromophore (phycocyanobilin)
which is essential for the green and red light receptor system.
Sequencing confirmed that the part is correct in the pSB1C3 vector.
We've Also Characterized the Following Parts
[http://partsregistry.org/Part:BBa_K608002 BBa_K608002] strong Promoter and strong RBS
[http://partsregistry.org/Part:BBa_K608003 BBa_K608003] strong Promoter , medium RBS
[http://partsregistry.org/Part:BBa_K608004 BBa_K608004] strong Promoter , weak RBS
[http://partsregistry.org/Part:BBa_K608005 BBa_K608005] medium Promoter , strong RBS
[http://partsregistry.org/Part:BBa_K608006 BBa_K608006] medium Promoter , medium RBS
[http://partsregistry.org/Part:BBa_K608007 BBa_K608007] medium Promoter , weak RBS
To quantify the strength of our PR-constructs we cloned GFP and RFP behind it
and quantified the protein output.
[http://partsregistry.org/Part:BBa_K608008 BBa_K608008]
constitutive strong promoter with medium RBS and GFP
[http://partsregistry.org/Part:BBa_K608009 BBa_K608009] Strong promoter with weak RBS and GFP
[http://partsregistry.org/Part:BBa_K608010 BBa_K608010] Medium promoter with strong RBS and GFP
[http://partsregistry.org/Part:BBa_K608011 BBa_K608011] Medium promoter with medium RBS and GFP
[http://partsregistry.org/Part:BBa_K608012 BBa_K608012] Medium promoter with weak RBS and GFP
[http://partsregistry.org/Part:BBa_K608013 BBa_K608013] strong Promoter with strong RBS and RFP
[http://partsregistry.org/Part:BBa_K608014 BBa_K608014 ] PR with RFP
[http://partsregistry.org/Part:BBa_K608015 BBa_K608015] PR with RFP
[http://partsregistry.org/Part:BBa_K608016 BBa_K608016] PR with RFP
[http://partsregistry.org/Part:BBa_K608017 BBa_K608017] PR with RFP
[http://partsregistry.org/Part:BBa_K608018 BBa_K608018] PR with RFP
 "
"
 Contact
Contact