Team:Freiburg/Modelling
From 2011.igem.org

Modelling: Rational protein design
The Idea
First there was the idea to replace the chemically produced Ni-NTA columns by some biological, reproducible material. It seemed unreasonable to produce an ion precipitating substance in a complicated, energy consuming way – completely artificial, while so many proteins in nature are capable of the same task – and much more complicated things. But how to harness that?
The second milestone in the generation of the idea came up in a lecture of Prof. Martin, about Nickel allergy. He found a mutation in the TLR-4 receptor that indroduces Histidines in the LRR motif of the protein, which are capable of binding nickel and thus forming complexes of receptors on the cell surface of immune cells, triggering the inflammatory reaction.
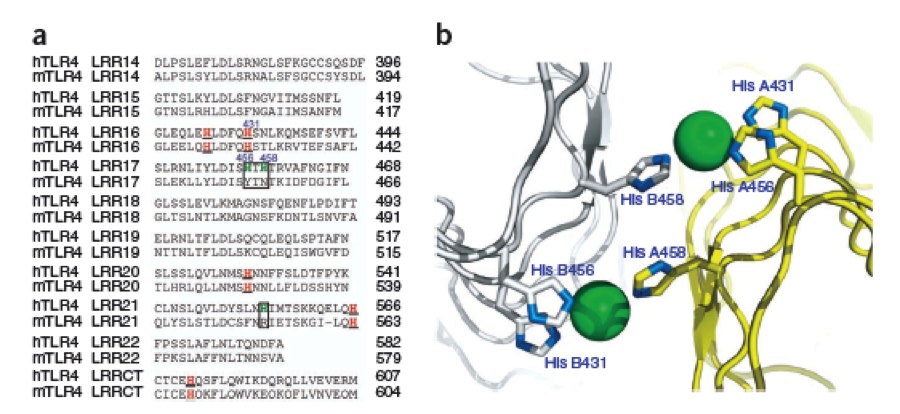 This was the necessary clue that gave answer to the question.
In the following work, a lot of investigation about the perfect Nickel precipitating protein was done. It seemed that there is no optimal natural protein which would serves our needs.
Furthermore, many of the natural LRR proteins – of course – have their own biological function, we we were afraid could interfere in the cellular system we wanted to express them in. Luckily there are over a thousand already known structures of LRR proteins, what gave us a broad choice of motifs to choose and compare.
The Construction of a new protein
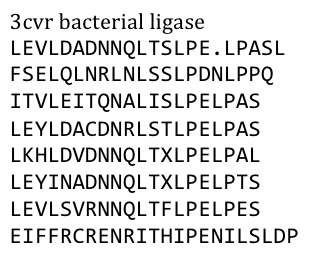
After a long and detailed search, we found out that it is most reasonable to use bacterial LRR motifs, since they seemed very well conserved in sequence, and they are the shortest – with only 21 aminoacids per LRR repeat. (Wei 2008, Kajava 1998). We wanted to have the protein as simple and as predictable in behavior and structure as possible.
Several search inquiries led to the most conserved and shortest of all bacterial LRR (PDB: NO), unluckily this protein is a toxin derived from Yersinia pestis. We of course did not want to mess around with toxins, especially not with one of the black plague. We also did not know what amino acid pattern on the LRR motif would cause the toxic effect. There probably would have been no harm from this protein alone, even if we did clone and express it, but why should we do that when we have a huge choice for free at hand anyway. Finally we settled on the bacterial ligase PDB:3CVR. A ligase would be definitively harmless, especially since we planned to restructure it completely.
This was the necessary clue that gave answer to the question.
In the following work, a lot of investigation about the perfect Nickel precipitating protein was done. It seemed that there is no optimal natural protein which would serves our needs.
Furthermore, many of the natural LRR proteins – of course – have their own biological function, we we were afraid could interfere in the cellular system we wanted to express them in. Luckily there are over a thousand already known structures of LRR proteins, what gave us a broad choice of motifs to choose and compare.
The Construction of a new protein
After a long and detailed search, we found out that it is most reasonable to use bacterial LRR motifs, since they seemed very well conserved in sequence, and they are the shortest – with only 21 aminoacids per LRR repeat. (Wei 2008, Kajava 1998). We wanted to have the protein as simple and as predictable in behavior and structure as possible.
Several search inquiries led to the most conserved and shortest of all bacterial LRR (PDB: NO), unluckily this protein is a toxin derived from Yersinia pestis. We of course did not want to mess around with toxins, especially not with one of the black plague. We also did not know what amino acid pattern on the LRR motif would cause the toxic effect. There probably would have been no harm from this protein alone, even if we did clone and express it, but why should we do that when we have a huge choice for free at hand anyway. Finally we settled on the bacterial ligase PDB:3CVR. A ligase would be definitively harmless, especially since we planned to restructure it completely.
To do this we needed to understand how the structure of the Ligase was made. It was that our desired protein LRR motif was only a part of a bigger protein “factory” which included several domains that seemed to serve distinct functions – which we did not want. So we took only the LRR domain. It looked to us, as if this domain had mostly a structural role in this “factory”, but we could not tell that exactly. To avoid any unspecific biological function it was necessary to rearrange the amino acid composition. We fed the protein sequence in a free online consensus sequence generator called weblogo from the Berkeley University.
 "
"
 Contact
Contact