Team:Freiburg/Description
From 2011.igem.org

Contents |
Precipitator
The Concept
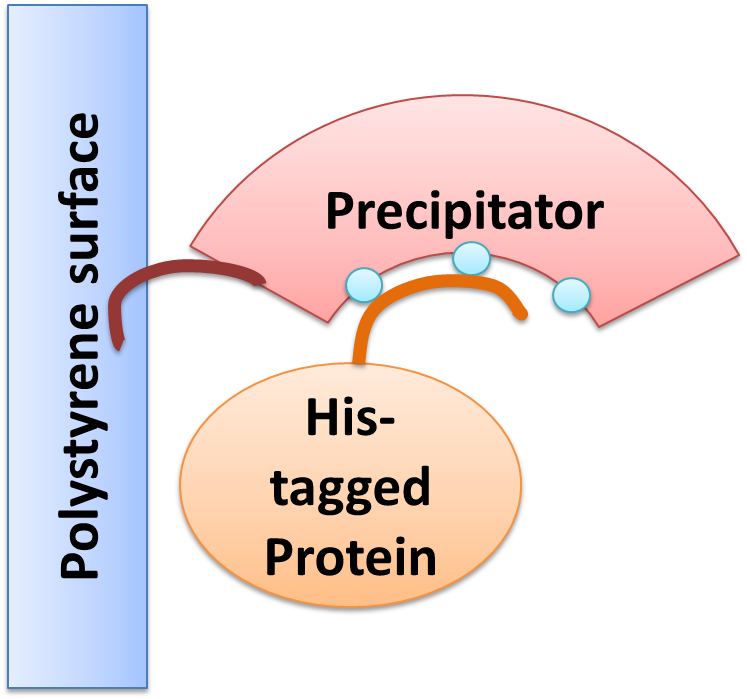
We created a cellular, self-replicating purification device for His-tagged proteins. It is a completely artificial fusion protein, which consists of a repeating LRRNT motif domain, coordinating Ni2+ Ions on its surface capped on N and C terminal ends by a hagfish sequence of a similar LRRNT motif. We named this construct “THE PRECIPITATOR”. A second domain, a short hydrophobic peptide stretch, binds a polystyrene surface, called the plastic binding domain. After expression of the Precipitator in a light inducible E. coli strain, the cells are lysed and the lysate is taken up with a serological pipette, in preparation of the actual protein purification steps. The underlying mechanism is comparable to Ni-NTA columns. Our Precipitator protein binds on the surface of the pipette, presenting the chelated Nickel ions. Free coordination sites of the Nickel ions are exposed, so that a His-tagged protein can attach to them. Cells expressing a His-tagged protein can be dissolved by the heat inducible lysis-device. Subsequently, when the lysate is taken up with a serological pipette coated with the Precipitator protein, the His-tagged proteins bind to it. Cell debris is then washed off, while the His-tagged protein stays and is eluted afterwards, in the same fashion as done in Ni-NTA columns with imidazole solutions, increasing in concentration. The His-tagged protein is finally captured in a distinct fraction.
Part design
The Precipitator BBa_K608406 protein is made of an artificial Leucine Rich Repeat (LRR) as the middle part of our own design, capped by C and N-terminal hagfish domain fragments.. This part is one version of three different designed to bind nickel by histidines, grouped together pointing away from the horseshoe shaped protein. Please see modeling for more details
Bacterial LRR Consensus of the central LRR fragment:
LxxLxLxxNxLxxLPxxLPxx
| Protein sequence CPSRCSCSGTEIRCNSKGLTSVPTGIPSS LTQLTKSNNHLHSLPDNLPAS LEVLDVSNNHLHSLPDNLPAS LEVLDVSNNHLHSLPDNLPAS LEVLDVSNNHLHSLPDNLPAS LEVLDVSNNHLHSLPDNLPAS LEVLDVSNNHLHSLPDNLPAS LKELALDTNQLKSVPDGIFDR LTSLQKIWLHTNPWDCSCPRIDY LSRWLNKNSQKEQGSAKCSGSGKPVRSIICP |
|
|
Colorcode (same in the sequence and video) | |
This protein can be used to complex Nickel or Cobalt. Histidines are positioned in such a way, that they can coordinate the ions from two to four orthogonal oriented directions. Free binding sites of the ions are then exposed, so that a His-tagged protein can attach to them. This protein can be used to complex up to 4 Nickel or Cobalt. The underying design of the protein is of a particular interest, too. LRR are highly conserved motifs throughout evolution. They appear in all kingdoms of life in almost every thinkable role (Ligases, Receptors, Toxins etc.). Their core is highly conserved and provides a very stable backbone, while the non-conserved aminoacids are almost freely interchangeable. Here we investigated an optimal set of non-conserved aminoacids by analysing large sets of similar proteins and databases. You can use this piece of work as a template to design your own protein and give it any function you like, by simply interchanging aminoacids and fusing other domains on the N or C termini. To guarantee proper folding and to shield off the hydrophobic core, a well studied fragment of an LRR protein coming from hagfish was used. This efficiency of this technique was proven before.(REFERENZ). To find out the most likely folding, we designed many different protein sequences, trying out a variety of sets of non coding aminoacids for the LRR and submitted these to the I-TASSER structure prediction
We only submitted one of the three versions, to reduce redundancy in the registry. Please contact us for any questions.
Plastic binding domain
One of the issues of our project „lab in a cell“ was to use endogenous proteins produced by the cell itself for specific purification and hereby to replace expensive columns. The „precipitator“ designed by our team contains a protein binding domain which complexes nickel and thus enables the binding of His-tagged proteins. After cell lysis the “precipitator” is freely dissolved in the cell lysate. To be able to isolate the “precipitator”-His-tagged-protein-complex from the other cell components it has to be immobilized by another protein domain. The part we designed for this function is the so-called plastic binding domain.
During routine phage display of random peptide libraries, phages were found that bound directly to the plastic surface of the used plastic micro titer plates. The number of plastic binding phages obtained during the phage display experiments depended on the saturation of the plastic micro titer plates with target protein for the antibody-binding phages and could be reduced by the use of blocking proteins as BSA or non-fat milk. Plastic binding phages were resistant to washing steps with PBS alone as well as to PBS in combination with BSA or non-fat dry milk. It was shown that plastic binding phages were even more difficult to recover by acid elution than the “normal” antibody binding phages (Adey et al., 1995).
The binding strength of the plastic binding protein was best on polystyrene plastic surfaces and also observed on PVC-plates (Adey et al., 1995).
The mechanisms by which the phage surface proteins bind to plastic are not well understood. The plastic binding amino acid sequences showed no obvious sequence similarity but where generally enriched by Tyr and Trp residues and are completely devoid of Cys residues. It is possible that the binding comes off non-specific hydrophobic interactions due to partial denaturation of the protein (Cantanero et al., 1980) and potentially due to interactions of the Tyr and Trp residues with the aromatic moieties of the polystyrene plastic surface of micro titer plates.
As well as high hydrophobicity does not necessarily imply plastic binding quality (Menendez et al., 2005) the observed plastic binding phages showed hydrophobic peptide sequences on their surfaces. Expressing such hydrophobic proteins in our host organism E.coli can lead to problems with inclusion bodies or decreased vitality up to cell death.
During our experiments we couldn’t obtain any clones containing both the gene for the plastic binding protein and a constitutive promoter-RBS-construct. We finally succeeded in expressing the plastic binding domain using an inducible promoter. To test the plastic binding domain and get data for our modeling we hereby used an IPTG-inducible promoter. In our completed “Lab in a cell” model the plastic binding domain, as a part of the “precipitator” would be expressed by one of our light inducible promoters.
Green light receptor
Sometimes a regulated and coordinated gene expression and therefore protein production is needed.
We decided to use light-controlled gene expression,because light is everywhere and always available.
The green light receptor is a light-sensing system from the cyanobactrium Synechocystis sp. PCC6803.
It consists of three parts interacting with each other in order to start regulated gene expression.
These parts are the following:
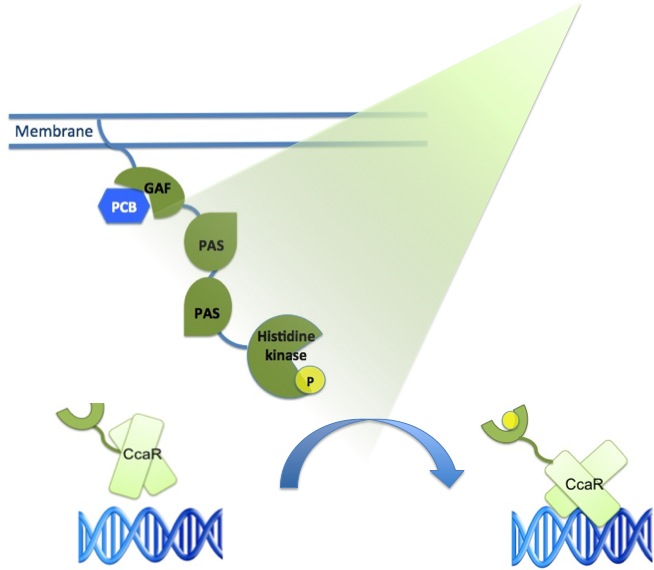
The main receptor is CcaS(1) ,a cyanobacteriochrome. It is made up of a N-terminal transmembrane helix, a cyanobactreria specific GAF domain, two PAS domains and a C-terminal histidine kinase.
To be fully functional CcaS has to bind the chromophore Phycocyanobilin (PCB) with its GAF-domain.
The GAF domain in this system has the ligation motif Cys-Leu, instead of the usual plant GAF-domain with Cys-His.
Its response regulator is CcaR (2), it belongs to the family of OmpR regulators. CcaR consists of an N-terminal receiver domain that can be phosphorylated by CcaS and a C-terminal DNA-binding domain that binds directly to the promoter region of cpcG2 (3). CpcG2 is an atypical phycobilisome which is playing a role in the energy transfer to photosystemI.
After light of a wavelength of 532 nm is sensed by the CcaS receptor, it changes its conformation. It undergoes autophosphorylation and the phosphate is transferred to the response regulator CcaR. Once phosphorylated, CcaR can bind to the specific promoter region of cpcG and activate gene expression.
As the green light sensing system was proven by J.J.Tabor to work also in e.coli, our plan is to integrate the genes for CcaS and CcaR into e.coli genome with a BAC (bacterial artificial chromosome). The gene which one wants to be regulated by green light has only to be inserted behind the cpcG2 promotor region and transferred into "our" e.coli strain.
[http://www.youtube.com/watch?v=SEz70ClCetM| Blue light] receptor
LOV- and tryptophan-activated protein (LovTAP)
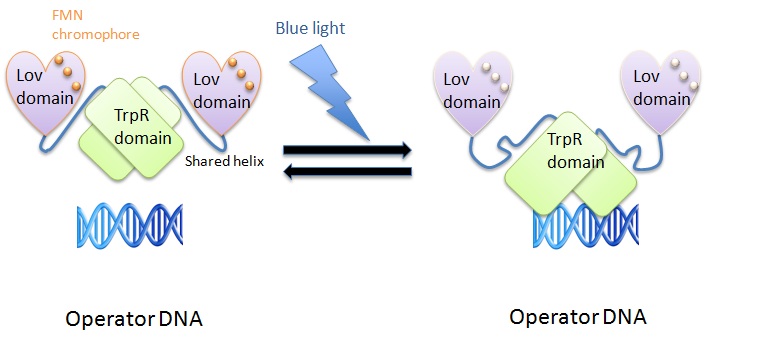
We used the LovTAP protein designed by Strickland et al. to induce protein expression using blue light (470nm). The LovTAP consists of the photoactive LOV2 domain of Avena sativa phototropin1 and the TrpR domain of E.coli as an output module. LOV2 (residues 404-543) is ligated via its carboxyl-terminal Jα-helix to a succession of 13 amino-terminal truncations of TrpR (residues 11-108).????
The shared helix acts as a rigid lever arm(which lever arm?) and the overlap(which overlap?) is most readily relieved by the disruption of contact between the shared helix and one domain or the other(what?). The helical contacts are integral to the structure of the domains, their disruption will cause a global shift in the conformational ensemble. Photoexcitation changes the conformational ensemble of the proteins and also changes the stability of the helix-domain contacts. This change shifts the relative affinity of the shared helix for each of the two domains. Photoexcitation sensed by one domain allows allosterical propagation to the other domain.
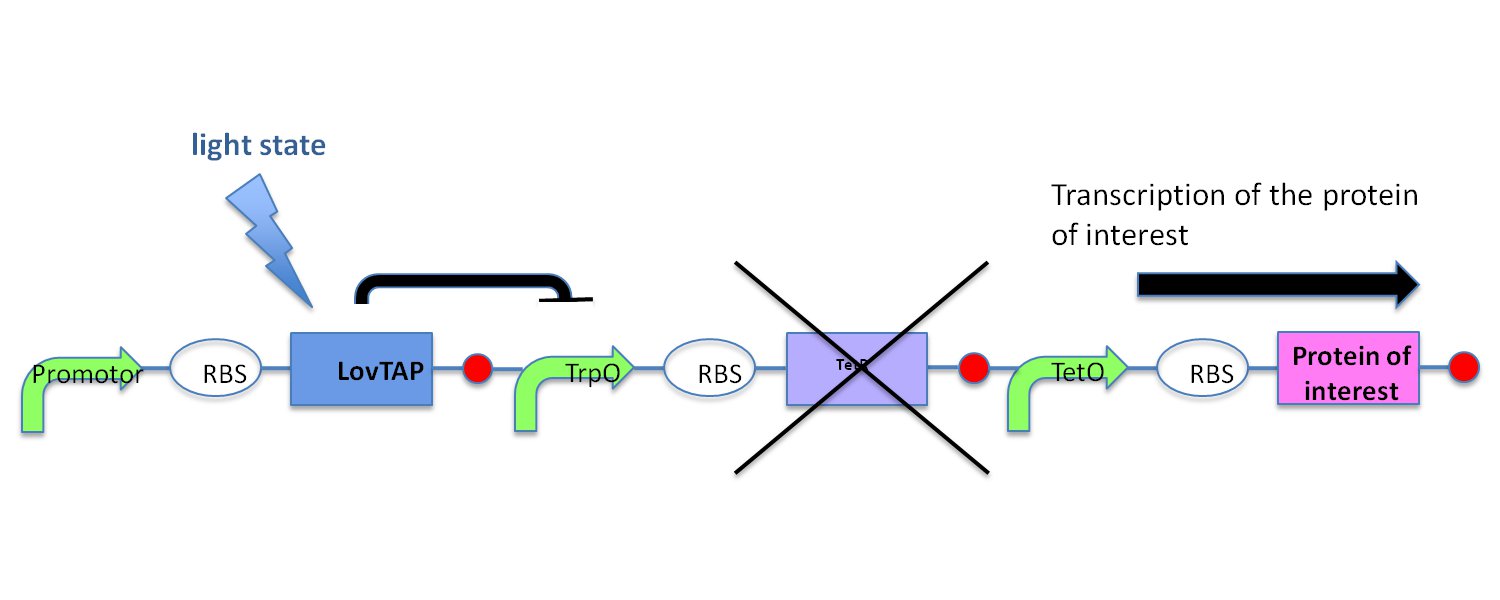
In the light state the LovTAP binds to the operator DNA and inhibits the gene expression. To avoid this inhibition, we cloned a Not-Gate after the LovTAP. Not-Gate is like an inverter, because it changes the input to its opposite. The Not-Gate consists of a tetracycline repressor and a TetR repressible promotor . In the light state the LovTAP binds to operators DNA and inhibits the expression of tetracycline repressor and the TetR promotor is active. In the dark state the tryptophan repressor is expressed and inhibits the expression of the protein of interest.
Red light receptor
Lysis cassette
The Concept
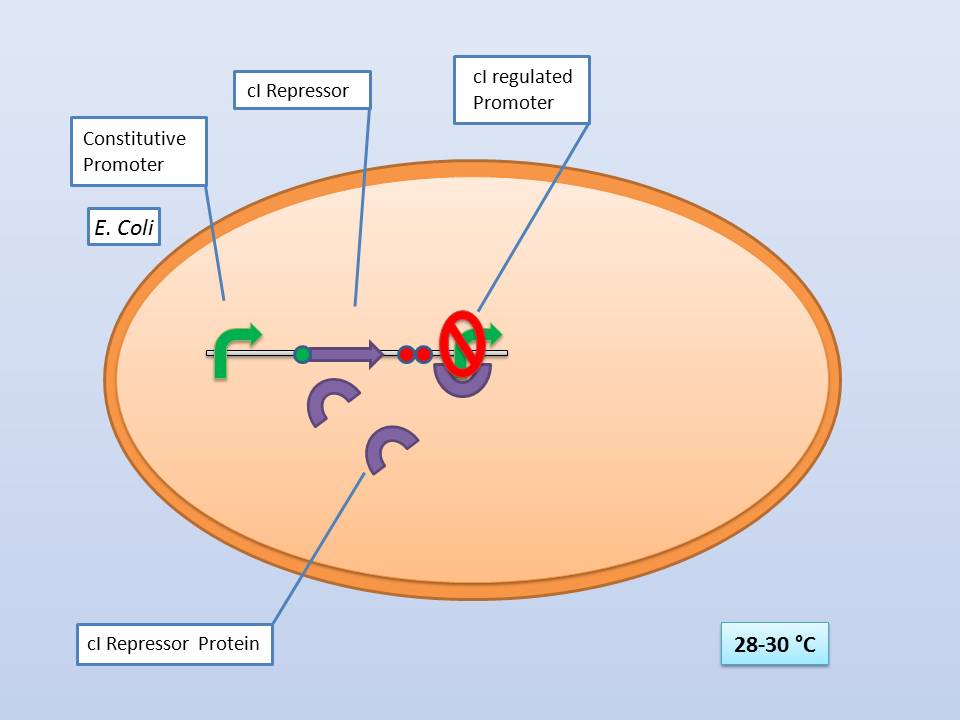
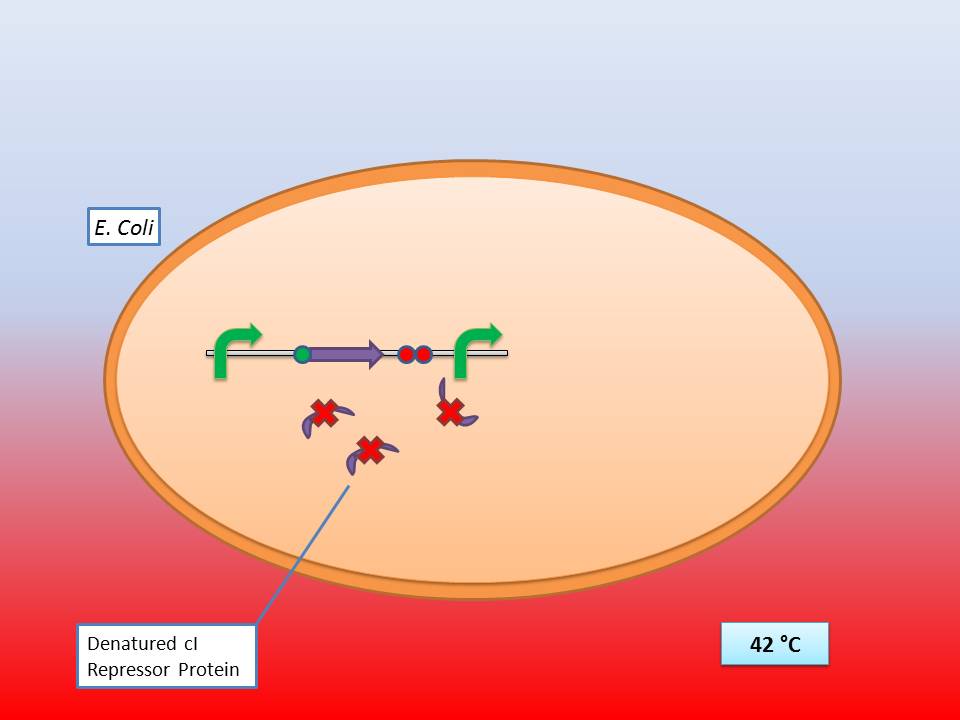
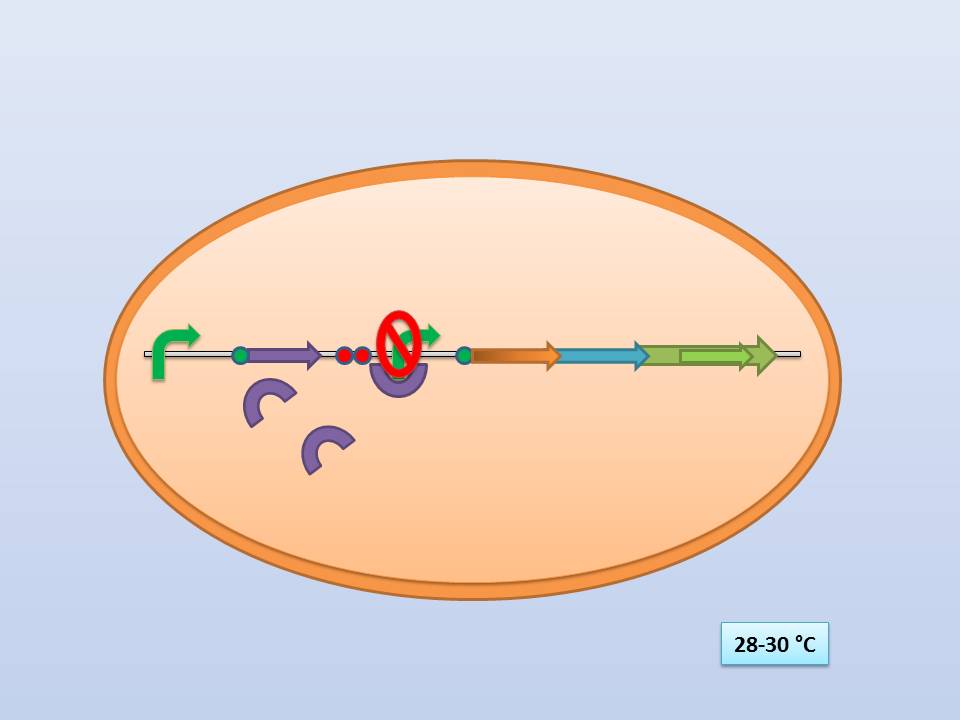
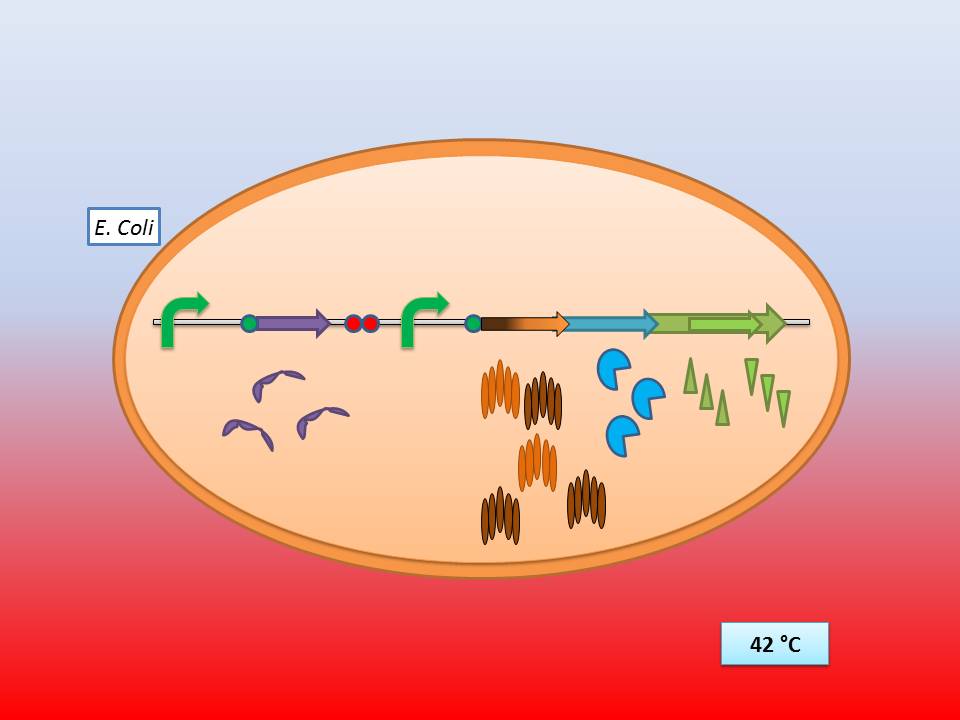
The idea behind our lysis cassette was to be able to achieve cell lysis by simply heating the bacteria at 42°C for a short period of time. This was to be accomplished by combining the biobricks BBa_K098995 (temperature sensitive promoter) and BBa_K124017 (Bacteriophage λ lysis genes). Sequencing of these parts (carried out by GATC Biotech GmbH) showed some fundamental inconsistencies leading to a lot of time spent on trying to fix them. This led to us not being able to send the whole planned lysis cassette to the registry on time, or even be able to test it correctly.
- Though some preliminary results showed that it probably works!*
Part Design
The temperature sensitive promoter (BBa_K608351)
It consists of a constitutive promoter (BBa_J23114), the temperature sensitive λ cI repressor protein which has a cI857 mutation that results in denaturation of the repressor when the temperature is raised from 30 to 42°C, and a cI regulated promoter based on the λ pR promoter (BBa_R0051).
- Troubleshooting: http://partsregistry.org/Part:BBa_K098995:Experience
|
|
|
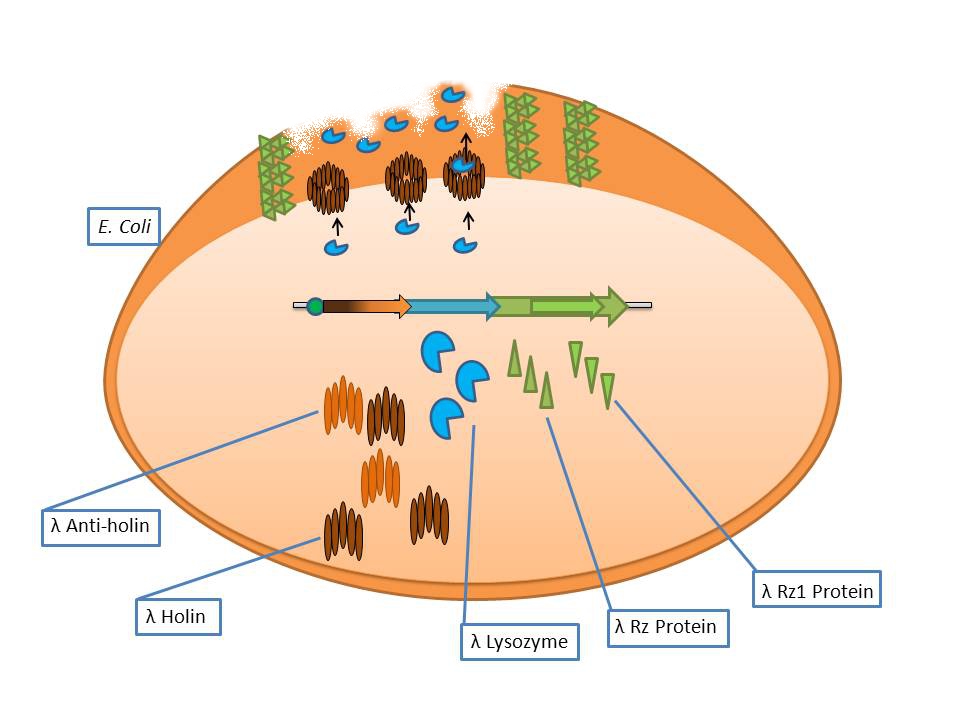
The λ lysis genes (BBa_K608352)
It consists of the standard bacteriophage λ lysis genes, ie S/S’, R, Rz/Rz1, coded in overlapping sequences with phase 1 and 2 frameshifts.
- S: λ Anti-Holin
- S’: λ Holin
- R: λ Endolysin
- Rz: Putative type-II signal anchor protein
- Rz1: Outer membrane lipoprotein
The phage λ lysozyme (R Endolysin) hydrolyses the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc), but in order to degrade the cell wall it requires a small transmembrane protein called holin which permeabilizes the membrane for endolysin to gain access to the murein [Ry Young et al]. The auxiliary lysis proteins Rz and Rz1 have long been known to play a vital role but have yet to be assigned a specific function stemming from functional and structural analysis. The assumption is that they form a complex spanning the periplasm and fuse the outer and inner membranes, removing the last physical barrier for cell lysis [Joel Berry et al].
- Troubleshooting: The biobrick did not have a RBS upstream of the CDS, so we had to clone BBa_B0034 in order to make a translational unit for cloning with the temperature sensitive promoter
The temperature sensitive lysis cassette
The much anticipated part that we frustratingly could not clone.
28-30°C incubation should repress the lysis genes, while a shift to 42°C would lead to their expression and finally cell lysis.
|
|
|
References
Nils B. Adey et al. 1995
“Characterization of phage that bind plastic from phage-displayed random peptide libraries”
Gene 156 (1995) 27-31
Alfredo Menendez & Jamie K. Scott 2005
“The nature of target-unrelated peptides recovered in the screening of phage-displayed random peptide libraries with antibodies”
Anal. Biochem. 336 (2005) 145-157
L. A. Cantarero et al. 1980
“The absorptive characteristics of proteins for polystyrene and their significance in solid phase immunoassays”
Anal. Biochem. 105 (1980) 375-382
Strickland et al. 2008
“Light-activated DNA binding in a designed allosteric protein”
National Academy of Sciences (2008) 10709-10714
Ry Young et al
“Phages will out: strategies of host cell lysis”
Trends in Microbiology Vol 8, Issue 3 (2000) 120-128
 "
"
 Contact
Contact