Team:Freiburg/Notebook/1 August
From 2011.igem.org

Contents |
green light receptor
Qickchange PCR of CcaS
Investigators:Julia
repeating experiment from___, because we had no positive transformation, probably chosen wrong annealing temperature before.
blue light receptor
PCR
Investigators: Sophie
As our last PCRs of the LovTAP with the Gibson overhangs didn't work well, we now try a PCR with primers without overhangs for Gibson assembly.
| Name: Sophie
| Date: 01.08.11 |
| Continue from Experiment: PCR (Date): 27.07.11
(Name): Sandra, Sophie | |
| Project Name: Blue Light | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | P35 |
| 2.5µl | Primer dw | P36 |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | M35 (LovTAP) |
| 0.5 µl | Phusion (add in the end) |
What program do you use?
LovTAP ohne Ueberhaenge
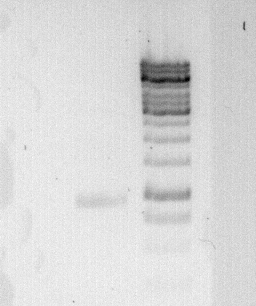
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
Labelled: M35 short; stored in Gibson Stuff box.
red light receptor
3A-assembly with pcyA and the terminator BBa_1006
Investigators:Julia
1.Digestion:
terminator (S21a), 26µl of the miniprep+ 12,1 µl H2O, cut with XbaI&PstI
pcyA (part BBa_I15009), 3 µl of miniprep + 35µl H2O, cut with EcoRI&SpeI
Vector-backbone (10 µl) pSB1T3, cut with EcoRI,PstI & DpnI
to each reation 5µl BSA(10x) and 5µl NEB buffer4 were added, digested at 37°C for two hours, inactivated at 80°C for 20min.
2.Ligation
2µl of digested pcyA,terminator and vector were added to 11µl H2O,2µl ligase buffer and 1µl T4 ligase.
Incubated at room temperature for 35 min, inactivated at 80°C for 20 min.
3.Transformation
Lysis cassette
Phage Lysis Cassette (K124017) + RBS (B0034)
Investigators: Theo
Digestion of 3A Assembly
| Name: Theo | Date: 01.08.2011 |
| Continue from Experiment : - | |
| Project Name: Phage Lysis Cassette (K124017) + RBS (B0034) | |
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng) (Vector:Insert ratio 1:3 in following Ligation)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Restriction enzymes you need to cut the vector, insert1 and insert 2:
| Components | | | |||
| DNA (500ng) | 10 | 9 | 8 | ||
| BSA (10x) (5μl) | |||||
| NEB4 Buffer (5μl) | |||||
| Enzyme 1 (1μl) | EcoRI + DpnI | EcoRI | XbaI | ||
| Enzyme 2 (1μl) | PstI | SpeI | PstI | ||
| H2O (38 μl- DNA) | 27 | 29 | 30 | ||
| In total 50 μl | |||||
Documentation:
Why are you doing this experiment? Where are the samples stored? Antibiotica resistance, vector used etc.
| A RBS (ribosome binding site) between the lamba cI regulated Promotor (R0051 of K098995) and the holin ATG of K124017 is absent. So a 3A assembly had to be performed in order to get a functioning Lysis cassette.
Resistance of RBS (B0034)=Cm and of Phage Lysis Cassette (K124017)=Amp, so the Vector is Tet (pSB1T3) |
Ligation
| Name: Theo | Date: 01.08.2011 |
| Continue from: 01.08.2011 Phage Lysis Cassette (K124017) + RBS (B0034) Digestion | |
| Project Name: Phage Lysis Cassette (K124017) + RBS (B0034) Ligation | |
Procedure
PCR tube:
total volume 20 μl
- add H2O (17 μl -X-Y-Z)
- add 2 μl Ligase Buffer 10x
- add Insert 1, Insert 2(when proceeding from 3A digestion use 2 μl of each)
- add Vector (20ng needed. When proceeding from 3A digestion use 2 μl)
- Add 1 μl T4-DNA Ligase
- Incubate 10-30 min at room temperature
- heat for 20 minutes at 80°C
- store at -20°C or directly proceed to transformation
| Name of part | Ratio Insert:Vector
= 3:1 or 1:1 | Volume (μl) | |
| X insert 1 | B0034 | 3:1 (1*3) | 3 |
| Y insert 2 | K124017 | 3:1 (2*3) | 6 |
| Z vector | pSB1T3 | 1:3 (1) | 1 |
| H2O | 7 |
Documentation:
Why are you doing this experiment? Where are your parts stored? Name the parts for ligation etc.
| Ligation step of 3A assembly.
Length of pSB1T3= ca 2200bp Length of K124017+Vector= ca 4500bp 4500/2200= ca 2 So 2*3 µl (since 3:1 is needed) = 6 µl
|
Transformation
| Name: Theo | Date: 01.08.2011 |
| Continue from : 01.08.2011 Phage Lysis Cassette (K124017) + RBS (B0034) Ligation
| |
| Project Name: Phage Lysis Cassette (K124017) + RBS (B0034) | |
Procedure
- take cells from -80°C freezer and put them on ice! (every eppi contains about 400 μl cells)
- thaw cells on ice 20 minutes
- pipette 50 μl cells and 2 μl DNA into eppi still on ice!
- Incubate for 30 minutes on ice
- Heat at 42°C for 60 sec
- Incubate on ice for 5 minutes
- Add 200 μl LB Broth
- Incubate for 2 hours at 37°C (cells with lysis cassette at 30°C!!)
- Plate 50 μl and 200μl on two different LB/Agar plates with appropriate antibiotic resistance
Documentation:
Why are you doing this experiment? Name of the sample? Where are they stored? Name the vector with inserts, antibiotika resistance etc.
| New Composite Part in pSB1T3 Vector with modified K124017 containing a strong RBS in front of first ATG |
Precipitator
PCR
| Name: Sophie
| Date: 01.08.11 |
| Continue from Experiment: replay of PCR 19.07.11 (Date) 19.07.11
(Name) Ruediger | |
| Project Name: GFP Pbd | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | P28 |
| 2.5µl | Primer dw | P18 / P19/ P20 |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | S 14 (GFP) |
| 0.5 µl | Phusion (add in the end) |
What program do you use?
Ruediger PCR (modified by Tobi and me. First 10 cycles with 55°C, next cycles with 62°C annealing temperature.
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
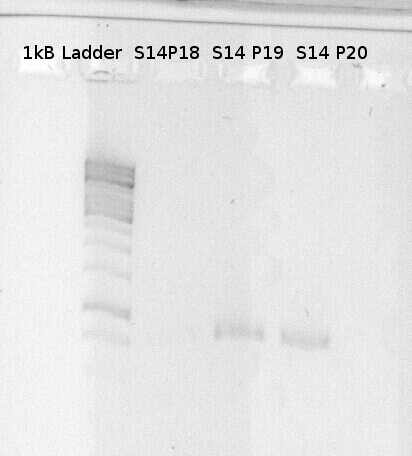
How did you label the PCR-Product, where is it stored and what do you do next?
 "
"
 Contact
Contact