Team:Freiburg/Notebook/1 September
From 2011.igem.org

Contents |
green light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
blue light receptor
Picking clones
Investigators: Sandra
There were several clones on the cm plates. Clones were picked and incubated in LB cm medium at 37°C.
Gibson primer design
Investigators: Sophie, Sandra, Ruediger
we designed some new gibson primers for the assembly of NOT and LOV-tap in a vector because we are not sure if we can achieve this by 3A assemblies.
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
3A Assembly of Quickchange-modified K098995 + modified Lysis genes K124017
Investigators:Theo
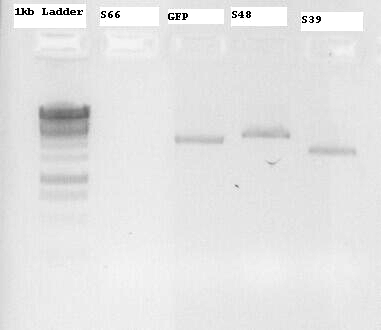
Another miniprep of the parts was done
- S66 = modified Lysis genes K124017 in pSB1A2
- GFP = E0040 in pSB1A2
- S48 = Quickchange-modified K098995 in pSB1A2
- S39 = Promotor (J23104) + RBS (B0034) in pSB1C3
As we thought, there is no band for S66, although there is DNA isolated from the mini-prep. It is probably a matter of making a stock from the false mini-prep. More answers on Monday or Tuesday
Precipitator
Ligation
| Name: Sophie | Date: 1.9.11 |
| Continue from Date: 31.08.11 Name: Sophie
Experiment: Digestion | |
| Project Name: | |
Procedure
PCR tube:
total volume 20 μl
- add H2O (17 μl -X-Y-Z)
- add 2 μl Ligase Buffer 10x
- add Insert 1, Insert 2(when proceeding from 3A digestion use 2 μl of each)
- add Vector (20ng needed. When proceeding from 3A digestion use 2 μl)
- Add 1 μl T4-DNA Ligase
- Incubate 10-30 min at room temperature
- heat for 20 minutes at 80°C
- store at -20°C or directly proceed to transformation
| Name of part | Ratio Insert:Vector
= 3:1 or 1:1 | Volume (μl) | |
| X insert 1 | GFP-pbd-PCR a/b | both | |
| Y vector | pGEX | ||
| H2O |
Documentation:
Why are you doing this experiment? Where are your parts stored? Name the parts for ligation etc.
| Why? To get data about the plastic binding for modeling
stored in: “Minipreps, verdaut”-box ligation-products stored in: ligation box parts for ligation: GFP-pbd-PCR a/b, pGEX |
 "
"
 Contact
Contact