Team:BU Wellesley Software/Notebook/CaseyNotebook
From 2011.igem.org
This summer the Wellesley HCI Lab worked on re-implementing an old system, G-nome Surfer 2.0, new and improved and optimized for prokaryotic genomes. To make an application practical for researching prokaryotic genomes we had to create a number of new interaction styles for dealing with navigation of a circular chromosome. Working on the system I worked on the new chromosome wheel, the publications system, the new primer designer and the associated BLAST.
G-nome Surfer Pro
Implementation
G-Nome Surfer Pro is written in C# using the Microsoft presentation SDK for use on the Microsoft Surface. After much trial and error we re-implemented many parts of the system using data binding which allows a complete separation of state and logic. (more information about data binding can be found at[http://msdn.microsoft.com/en-us/library/ms752347.aspx MSDN]) By using data binding we have managed to create remarkably stable code and haven't seen any crashes at all in usability testing and user studies. However, the structure and time required to set up the system is difficult to manage and some components outside the core (like primer designer) are implemented without. Fortunately, the underlying structure allows for editing of individual components and collaboration through the svn repository.
Navigation
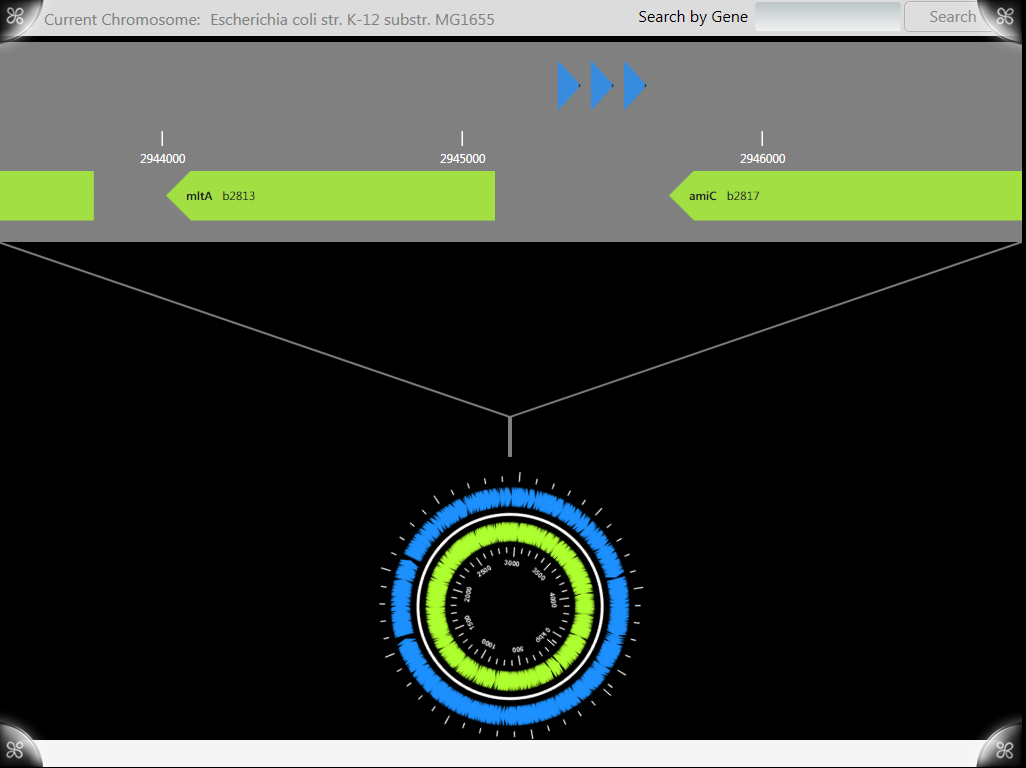
The central chromosome wheel went through several iterations before arriving at the final simple rotation model we currently use. An early implementation allowed for zooming through use of a pair of arms (like in the early paper prototype.) Like many of the early user controls the arms involved a fair amount of hacking through the presentation logic and item templates through the .xaml files. With the option of zooming through the hands the issue of scale became an obvious problem with the system. Based on the total number of base pairs in a chromosome it turns out that any reasonable zoom level is represented by fractions of a degree which made the use of the arms for zooming impractical.
In later versions the arms were abandoned in favor of a single line and a constant zoom level. The new wheel uses images generated by [http://wishart.biology.ualberta.ca/cgview/ cgview] and tracks position in the chromosome based on the orientation of the object.
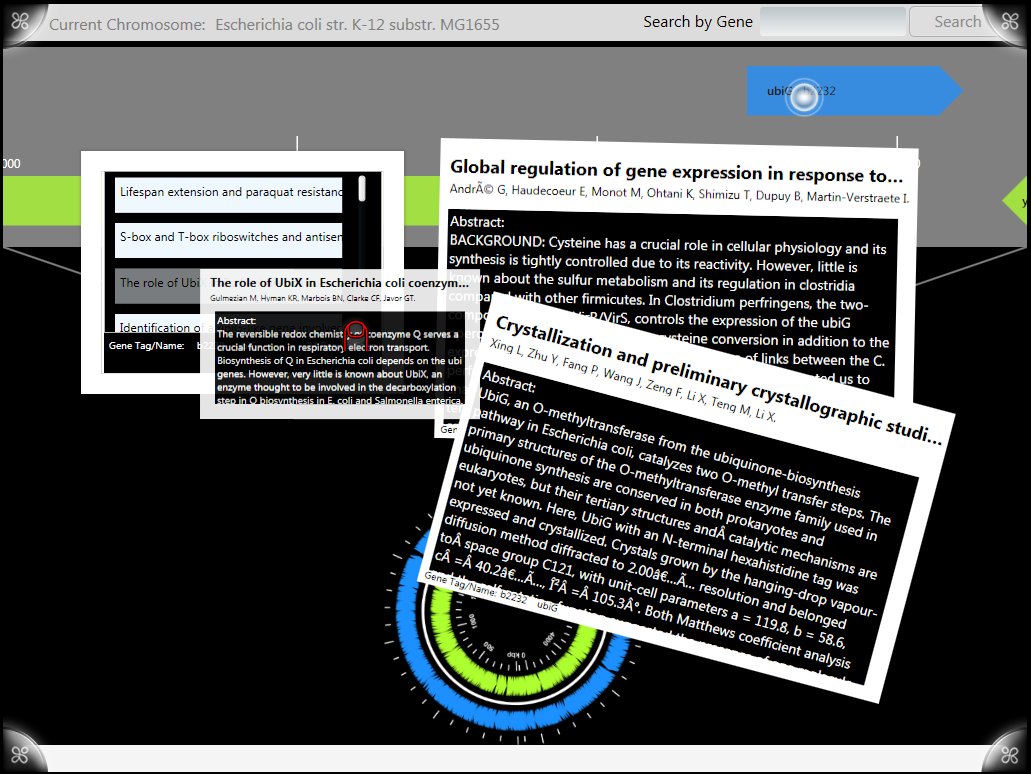
Publications
Another system that was hacked thoroughly in earlier implementations, the publications system interfaces with and pulls abstracts from [http://www.ncbi.nlm.nih.gov/pubmed PubMed.] Because implementing drag and drop is not a simple task with the current SDK, early implementations involved faking the interaction through snapping and dragging multiple objects with a single objects. Later implementations took advantage of data binding, item templating and the surfaceListBox object to create a more intuitive, stable drag and drop experience.
Subsystem: PrimerGenie
Primer Designer
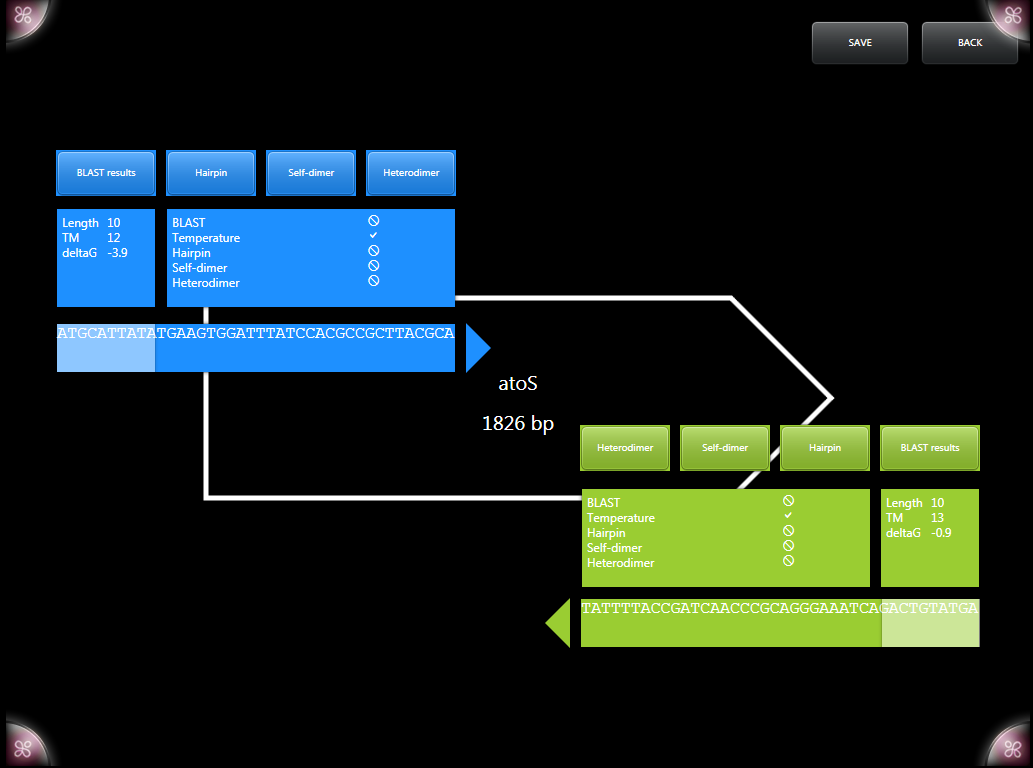
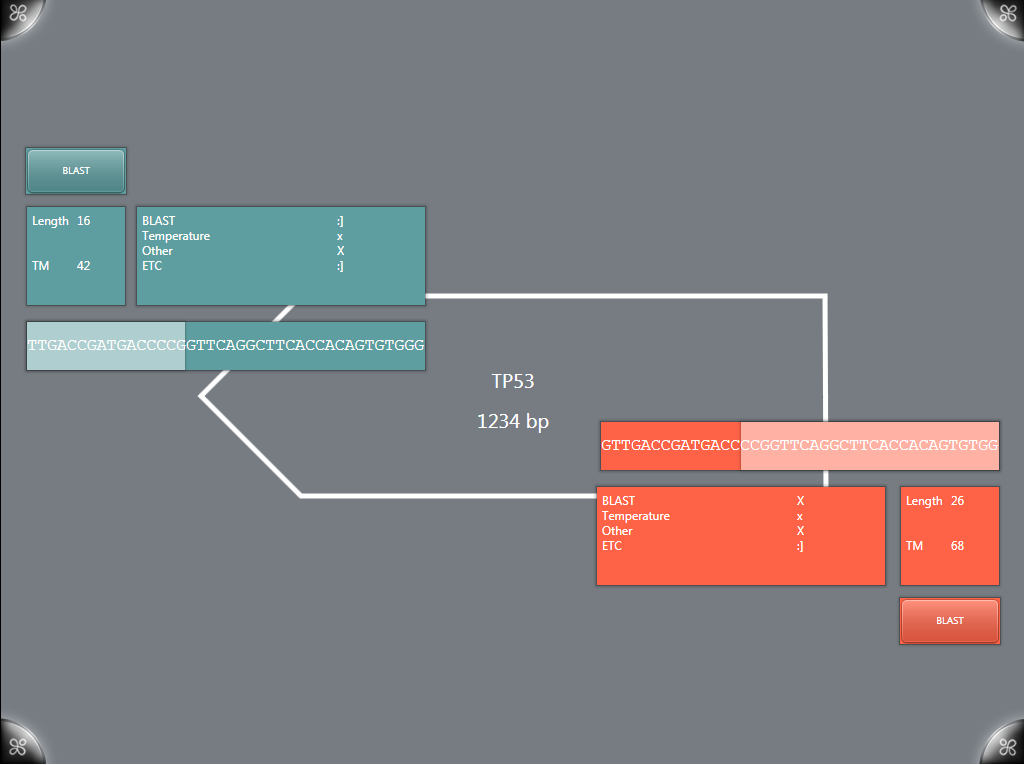
The primer designer was a completely new addition to the G-Nome Surfer system for IGem. Using the surface as a primer designer allows for a more streamlined and collaborative primer design experience. Users can track a number of stats that are updated as the selected sequence is updated including Gibbs free energy and the melting temperature of the primer. A number of tests can be performed within the designer such as testing for hairpins, heterodimers, self dimers using algorithms from the BU comp team and viewing of BLAST results. Cut sites from the standard biobrick restriction enzymes are automatically visualized within the primer; later versions may allow for the use of user selected restriction enzymes and cutsites.
BLAST visualization
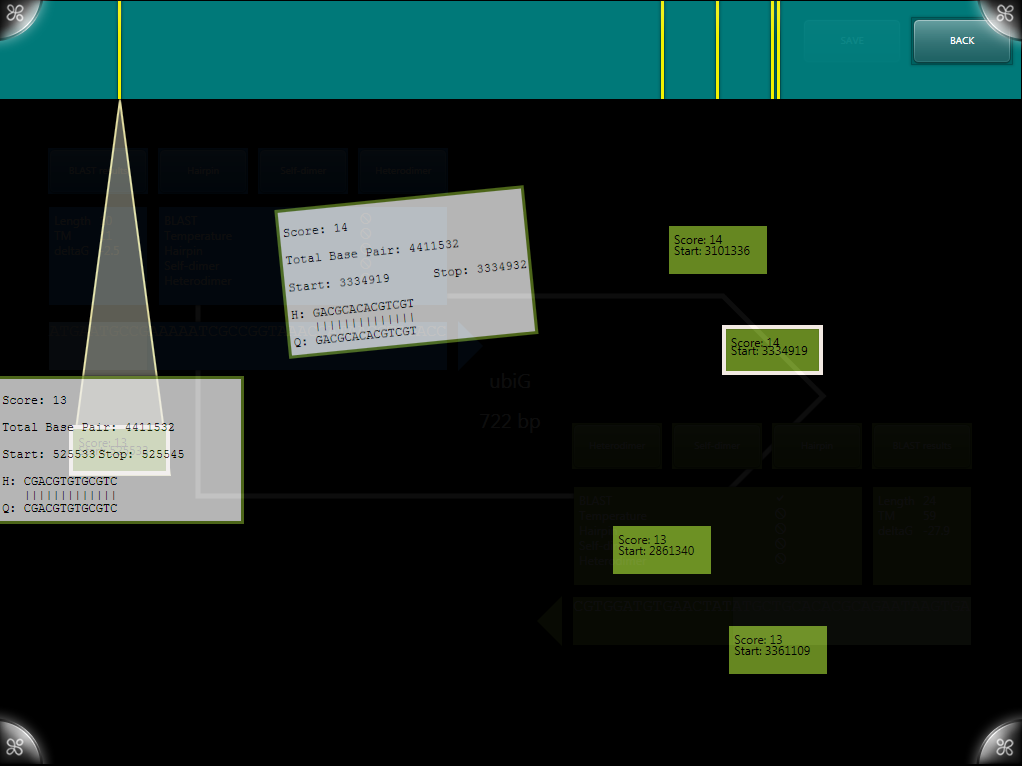
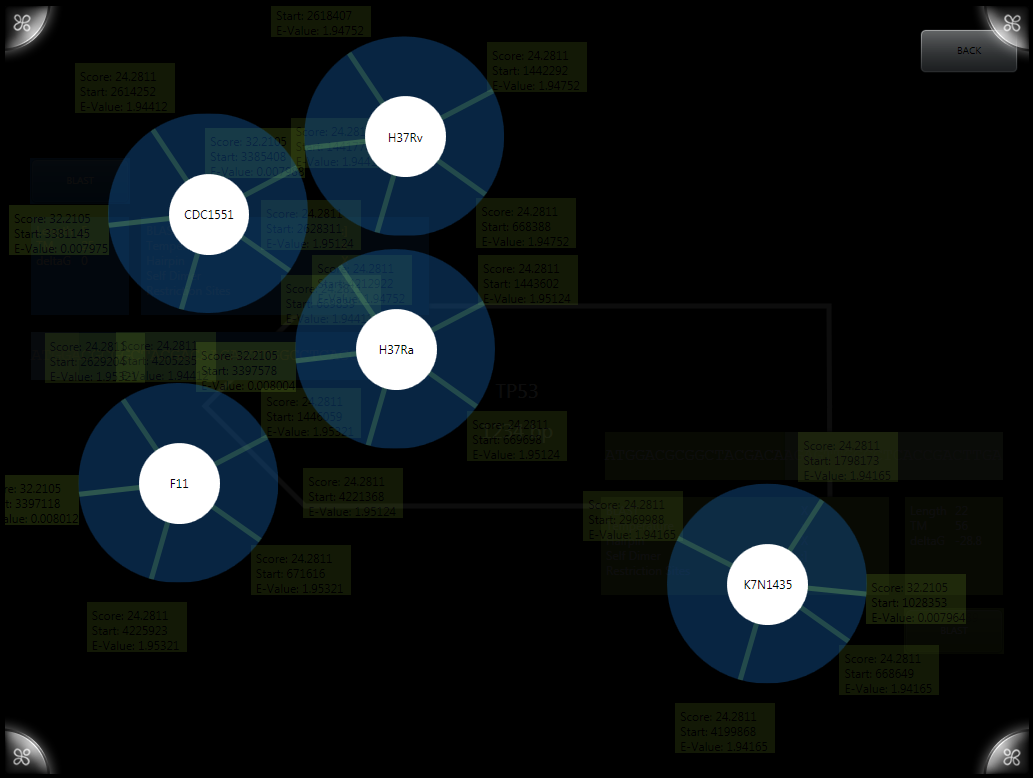
BLAST results are generated using the [http://blast.ncbi.nlm.nih.gov/Blast.cgi NCBI BLAST algorithms] and displayed on a layer above the primer designer. The original implementation allowed viewing of results compared to multiple genomes visualized as circles with results. Results can be expanded for more information about a given BLAST result.
A newer version simplifies the visualization down to a single genome (in our case, H37RV) and displays it as a single bar. Results are staggered and can again be expanded, now including more information including the alignment of the sequences.
Subroutine: Weekly
week of 7/11
Finally started work on the primer designer. The plan is to make the biobricks designed here eventually save (through use of a token) to the Clotho database. That's probably a while off for now though. Progress was pretty rapid, just getting the .xaml all set up and getting the interaction styles all set up.
week of 7/18
Moved on to the BLAST visualization for primer designer this week. Spent a lot of time just getting the geometry to work out though, because it turns out doubles sometimes round to zero. gross. We spent a lot of time getting the old BLAST .xml parser and new commandline interface playing nice with one another and the visualization. We also looked into some other tests- hairpins, selfdimers, heterodimers etc etc etc. Which meant we got to work more closely with the computational team at BU. We talked to Jenhan and he gave us his code for alignment to get the primer designer up and running in a real way.
week of 7/25 Finally starting to work on integrating PrimerGenie in with the rest of the G-Nome Surfer system. We had a long day of usability testing on the 27th and spent the rest of the week correcting for small usability issues. We also spent a little while hooking up the visualization for restriction sites for the biobrick enzymes only. Maybe in the future we'll allow a user to specify them? Not much time for that now though, with a user study with BU on Monday. Saturday afternoon was dedicated to getting threading set up so that the entire interface doesnt lock up when running BLAST.
week of 8/1 Last week on campus, spending time on documentation for the first time in a while. There is lots of data from the user study on Monday to go through and transcribe.
 "
"