Team:Peking S/modeling/summary
From 2011.igem.org
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Summary
Quantification makes synthetic biology different. Mathematical model and computer-aided design software enable synthetic biologists to implement synthetic gene networks with a rational guidance.
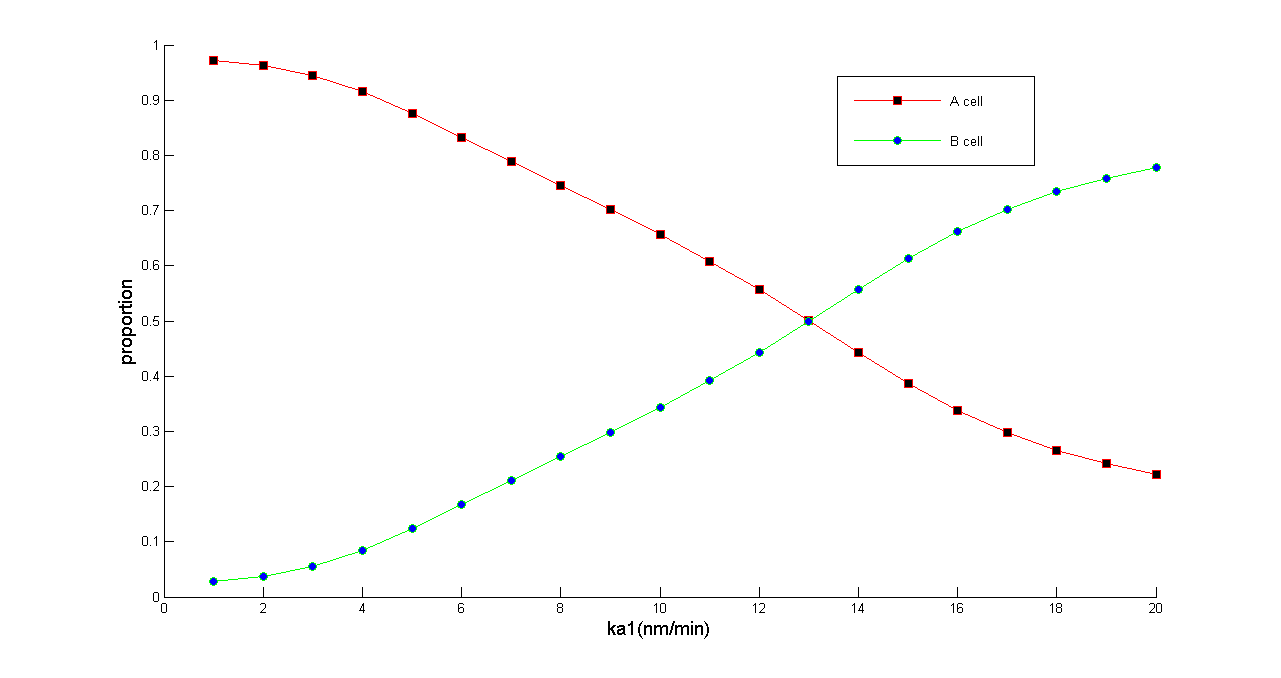
After designing genetic circuit and before experimental implementation in bench, we constructed a set of ordinary differential equations to mathematically analyze the population density balancer. Modeling results suggests that our population density balancer can control the population of A (B) cell in arbitrary proportion. And the whole system is robust enough to perform as population balancer with minimal possibility as an oscillator due to its topology. Under this topology, our system could perform as a balancer in a wide range of parameter space while the ratio of A/B cell can be still tuned among a wide range.
For more details, please click here.
We have harvested, re-designed and quantitatively characterized enough ‘chemical wires’ for developing this ‘chemical wire’ toolbox. But this should not the end. We noticed that there is actually a trade-off between signaling speed and layers of cell-cell signal transduction when distributing a logic function among a synthetic microbial consortium. To figure out how to cope with this trade-off, we next sought to propose design rules and thus to develop software to facilitate the distribution of Boolean logic gene network among a synthetic microbial consortium. By applying the principles to our sixteen two-input, one-output circuits discussed above, we discovered that all other logic circuits could be implemented within fewer than 2 cells.
Design and Experimental Results of XOR gate
For more details, please click here.
 "
"