Team:SJTU-BioX-Shanghai/Project/Subproject1
From 2011.igem.org
|
|
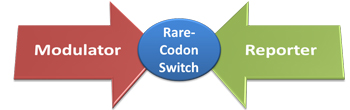
Rare-Codon SwitchCodon degeneration indicates that more than one codon can be used to encode an amino acid. Every species has codon usage bias. For one species, some codons are rarely used(less than 1%). These codons are called rare codons. Codon usage and tRNA amount are closely related. When heterogenous protein is expressed, differences in codon usage in mRNA may impede translation because one or more tRNAs are rare in the population, especially when there are multiple consecutive rare codons at the N-terminus of a coding sequence. This has proposed a new way to control protein expression on translational level. In E.coli, the rarest used codon is AGG for Arg. By adding AGG codons to the site close to initial codon and regulating the abundance of rare tRNA, we can regulate protein biosynthesis on translational level.
IntroductionWe design a Rare-Codon Switch controlling protein biosynthesis. We can control the translation process by controlling how well the ribosome can get through a tandem of rare codons in the target protein's mRNA. This process can be achieved by controlling
|
 "
"