Team:DTU-Denmark/Background sRNA
From 2011.igem.org
{:Team:DTU-Denmark/Templates/Standard_page_begin|Background}}
What is sRNA
In our project we dig into the fascinating world of sRNA regulation with the aim of engeneering a so-called trap sRNA regulating module and hereby creating a universal tool for a whole new way of regulating gene expression in synthetic biology. To fully understand our project, at basic knowledge of sRNAs, the way they regulate gene expression, the players involved (the RNA chaperone Hfq) and the unique features of the regulation is required. So continue reading if these words are new to you.
An recurring theme for sRNA regulation is the diversity of its mechanism; some sRNAs are repressors other activators, some act catalytically others non-catalytically, some require Hfq others not and so on. All in all this illustrates the extensive variety of sRNAs and how they have been tuned to operate in a range of regulatory niches in the cell during the process of evolution.
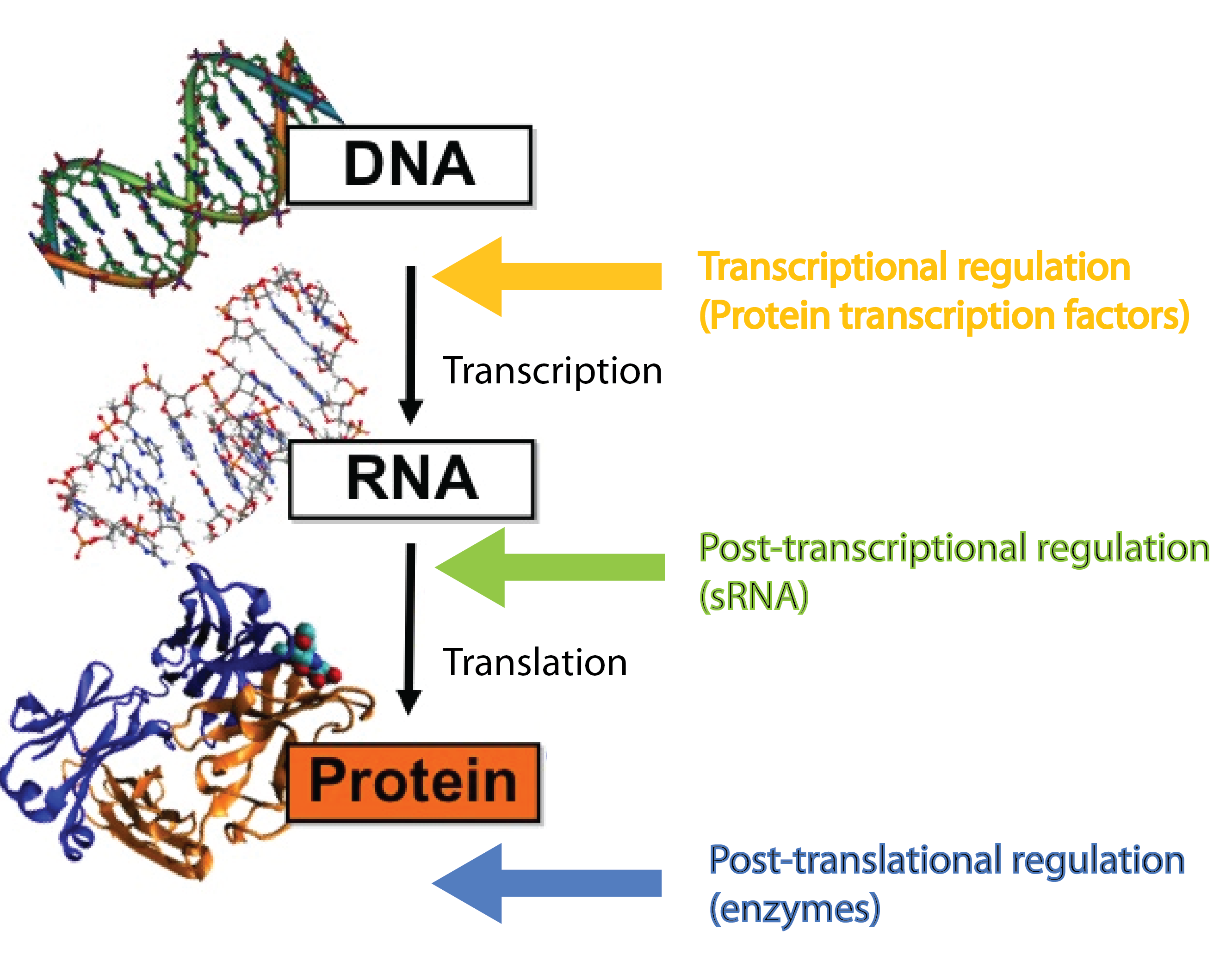
Many different kinds of RNA molecules are present in the cell; some of these are never translated into proteins. This group is collectively called non-coding RNAs, and includes, in addition to small RNA (sRNA), also rRNA and tRNA as well as more exotic RNA classes. Bacterial sRNAs are small RNA molecules (normally 50-200 nucleotides) which are extremely interesting as they have been demonstrated to play central regulatory roles in prokaryotes. There are two groups of sRNAs; the first group regulates gene expression by binding to proteins (e.g. transcription factors) and inhibiting their function, the second group regulates gene expression by binding to mRNAs. In our project we are focusing solely on the latter group, which regulate gene expression at the post-transcriptional level[1][2]. Whenever we mention sRNA we refer to those whit mRNA targets. Regulatory RNAs are not restricted to prokaryotes. Eukaryotes (including mammals) use micro RNA (miRNA) in post-transcriptional regulation, which is a functional analog to sRNA[3].
sRNAs act at the post-transcriptional level, a level complementary to the level at which protein regulators operate. Protein transcription factors operate before sRNAs at the transcriptional level whereas enzymes such as kinases and phosphatases act after sRNAs at the post-translational level[3]. Combining different layers of regulation enables interesting regulatory outcomes, eg. increase the number of genes regulated in response to a given signal or extremely tight repression of genes - sRNA can silence leaky promoters[3]. Transcription factors typically bind the target gene within ~100 nucleotides of the -10 and -35 region which set an upper limit for the number of transcription factors that can regulate a given gene. By targeting a complete different part of the gene, sRNAs can increase the number of signals a given gene can sense.
References
[1] Beisel, Chase L., and Gisela Storz. “Base pairing small RNAs and their roles in global regulatory networks.” FEMS Microbiology Reviews 34, no. 5 (2010): 866-882.
[2] Levine, Erel, Zhongge Zhang, Thomas Kuhlman, and Terence Hwa. “Quantitative characteristics of gene regulation by small RNA.” PLoS biology. 5, no. 9 (2007).
[3] Waters, Lauren S., and Gisela Storz. “Regulatory RNAs in Bacteria.” Cell 136, no. 4 (2009): 615-628.
 "
"