Team:Freiburg/Notebook/24 August
From 2011.igem.org

Contents |
green light receptor
Digest of CcaR
Investigators: Jakob
- 10µl BSA
- 10µl NEB
- 2µl DpnI
- 38µl DNA
Digest for 2h at 37°C
Transformation of CcaR
Investigators: Jakob
Testdigest of quickchanged CcaS
Investigators: Julia
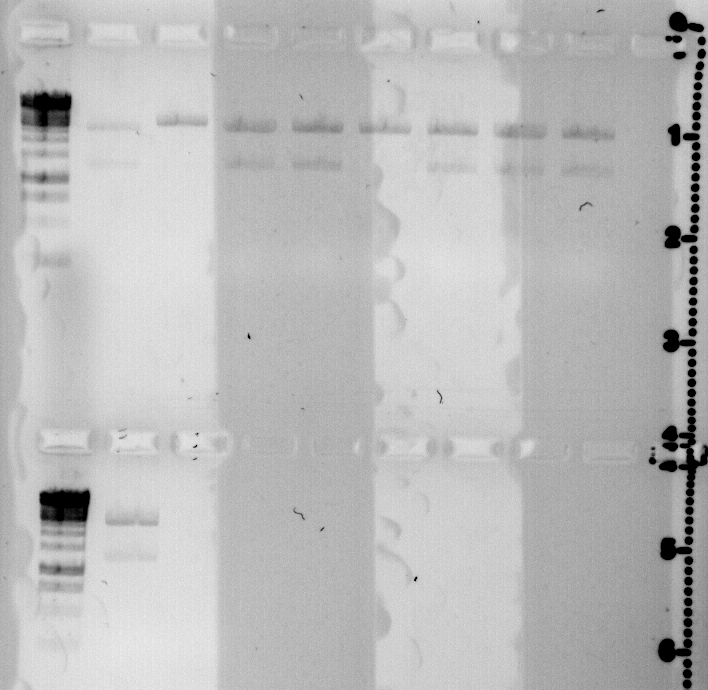
today I prepared 9 minipreps of the the quickchanged CcaS.
They were digested with EcoRI and loaded on a gel.
Those which are not cut twice were changed during PCR correctly.
We will send them for sequencing today.
gel loaded with: Ladder 1 kb, qS50 a,b,c,d , qS51 a, b, c, d
lower lane:Ladder 1 kb qS51 e
positive clones are qS50b and qS51a.
concentration of minpreps: qS50b: 88,6 ng/μl and qS51a: 81,1 ng/μl.
blue light receptor
Transformation
Investigators: Sandra
Repeat of the transformation from yesterday with the Lovtap-NotGate-pSB1A3 vector.
PCR
| Name: Sophie
| Date: 24.08.11 |
| Continue from Experiment: primer design (Date) 19.08.11
(Name) Sandra | |
| Project Name: Blue light | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | P24 |
| 2.5µl | Primer dw | P79 |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | M45 |
| 0.5 µl | Phusion (add in the end) |
What program do you use?
20 cycles with an annealing temperature of 65/68/71 ºC (gradient-PCR) and touchdown then 10 cycles with an annealing temperature of 65/70/75 ºC and touchdown
digested with DpnI
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
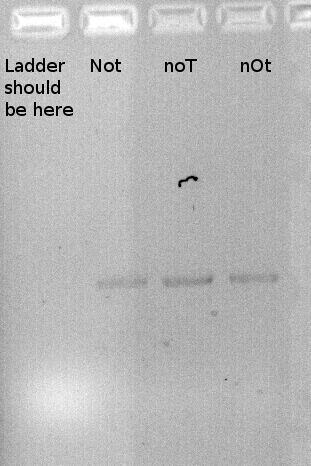 /border
/border
How did you label the PCR-Product, where is it stored and what do you do next?
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
NAME OF YOUR EXPERIMENT
Investigators: NAME
 "
"
 Contact
Contact