Team:Freiburg/Notebook/12 August
From 2011.igem.org

Contents |
Meeting
attendants: Jakob, Julia, Manuel, Sandra, Sophie, Theo, Tobias
Time: 10:00 - 11:30
green light receptor
already done:
- quick change with CcaR didn't work
- colonies for pcyA and ho1
To-do:
- order new primer for the quick change
- cph8: we will use the primer and protocol from the Upsalla team
- test if the colonies are positive
blue light receptor
already done:
- PCR to amplifiy the receptor with new primer pair
To-do:
- check if the PCR worked
red light receptor
already done:
To-do:
Lysis cassette
already done:
- lysis cassette with RBS
- colonies for the GFP-PBD
- inducible promotor + PR done
To-do:
- overnight culture of the GFP-PDB colonies, miniprep, test digest and sequencing
Precipitator
already done:
- waiting for the genes
To-do:
still more waiting to do
green light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
blue light receptor
Gel of the PCR product: LovTAP
Investigators: Sandra
To confirm the PCR worked this time, we loaded the sample onto a gel. The size of the fragment should be 1000bp. The gel showed a band at 1000bp. We digested the PCR product with DpnI and then purified the DNA with the PCR purification kit to further use the PCR product for 3A-assembly.
3A-assembly of Not-Gate, LovTAP in pSB1T3
Investigators: Sandra
Digestion
Amount of DNA and H20:
| Sample | DNA μl | H20 μl |
| LovTAP | 5 | 33 |
| Not-Gate | 2 | 36 |
| pSB1T3 | 20 | 18 |
Enzymes necessary for digestion:
| LovTAP | Not-Gate | vector | |
| enzyme 1 | EcoRI | XbaI | EcoRI |
| enzyme 2 | NheI | PstI | PstI |
- Incubation at 37°C for 6 hours
- 20 minutes at 80°C
red light receptor
Picking clones of pcyA and ho1
Investigators: Sandra
Picked 2 clones of each plate:
- 1pTd (PR1+pcyA+terminator)
- 2pTd (PR2+pcyA+terminator)
- 1pTb (PR1+pcyA+terminator)
- 2pTb (PR2+pcyA+terminator)
- 1L23 (PR1+ho1+terminator)
- 2L23 (PR2+ho1+terminator)
Incubation over night.
Lysis cassette
Miniprep
Investigators: Jakob, Ruediger, Theo
| Name: Jakob, Ruediger, Theo | Date: 12.08.2011 |
| Continue from Experiment: Transformation, 10.08.2011 | |
| Project Name:
Lysis systeme, plasticbinding, GST-Tag, IPTG inducible promotor, LOV-Tap | |
Documentation:
Why are you doing this experiment? Name the parts which you extract.
- Need the DNA concentration
Describe your results and mistakes and measure the DNA concentration with the Nanodrop and note the results. How did you label your probes and where are they stored?
| Sample ID | Nuclein Acid Conc. ng/µl | | |
| LOV 3A | 81,8 | 1,88 | 2,23 |
| PR4+GFP | 87,1 | 1,85 | 1,93 |
| PR6-1 | 124,7 | 1,89 | 2,19 |
| PR6-2 | 138,1 | 1,88 | 2,14 |
| PR6-3 | 67,2 | 1,94 | 2,09 |
| PR6-4 | 96,5 | 1,85 | 1,58 |
| PR6-5 | 102,7 | 1,94 | 2,14 |
| PR6-6 | 111,5 | 1,88 | 1,99 |
| PR6-7 | 40,1 | 1,99 | 1,94 |
| PR4-1 | 71,3 | 1,90 | 1,79 |
| PR4-2 | 119,6 | 1,83 | 1,69 |
| PR4-3 | 135,4 | 1,84 | 1,83 |
| PR4-4 | 99,2 | 1,86 | 1,85 |
| PR4-5 | 90,9 | 1,84 | 1,67 |
| Lys+RBS-1 | 33,1 | 1,99 | 1,87 |
| Lys+RBS-2 | 43,8 | 1,94 | 1,91 |
| Lys+RBS-3 | 27,7 | 1,98 | 1,82 |
| Lys+RBS-4 | 38,9 | 2,02 | 1,92 |
| Lys+RBS-5 | 31,0 | 2,03 | 1,92 |
| Lys+RBS-6 | 39,9 | 2,00 | 1,84 |
| PGEX6P1-a | 65,9 | 1,94 | 2,02 |
| PGEX6P1-b | 65,3 | 1,90 | 1,98 |
| PGEX6P1-c | 70,6 | 1,95 | 2,05 |
| PGEX6P1-d | 43,6 | 1,82 | 1,40 |
| PGEX6P1-e | 53,9 | 1,87 | 1,75 |
| S54 | 219,6 | 1,87 | 2,21 |
| S54 | 216,4 | 1,88 | 2,21 |
Precipitator
Cloning
Investigators: Sophie
continue from experiment: Miniprep (Jakob/Ruediger, 12.8., see lysis cassette)
project name: inducible promoter for Pbd
parts: S54 (Eco & Spe)
GFP-Pbd ε5 (Xba 7 Pst)
psb1t3 (Eco & Pst)
stored in: Ruediger's box
Ligation
Investigators: Sophie
continue from experiment: Cloning (today, me)
project name: inducible promoter for Pbd
parts: S54 (Eco & Spe)
GFP-Pbd ε5 (Xba 7 Pst)
psb1t3 (Eco & Pst)
name of ligation product: IPTG+Pbd
stored in: Ruediger's box
Testdigest
Investigators: Sophie
continue from experiment: Ligation (today, me)
project name: inducible promoter for Pbd
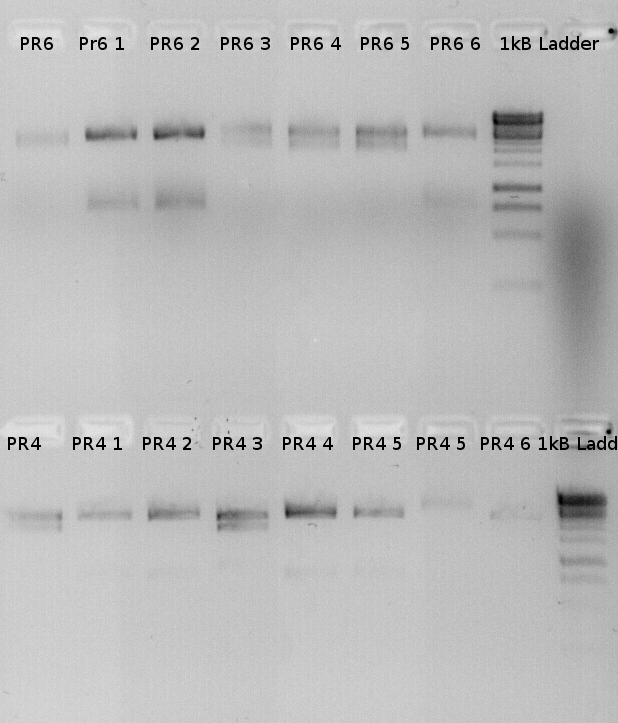 PR6 6 and Pr 4 4 were sequenced but PRs were missing again. Inducible promoter seems to be necessary.
PR6 6 and Pr 4 4 were sequenced but PRs were missing again. Inducible promoter seems to be necessary.
 "
"
 Contact
Contact 










