Team:BU Wellesley Software/Notebook/KelseyNotebook
From 2011.igem.org
Want to get updates about my work on iGEM and other HCI projects? Follow me on Twitter: KelseyTempel[1]
Week by Week
30 May-
Tuesday was my first day in the HCI lab. Some lab mates and myself went over some basic Microsoft Surface programming to get ready for the upcoming iGEM project.
Wednesday-Friday, and Monday, I went with the Wellesley team to BU for Boot Camp! I haven't taken a biology class since 8th grade so all the material seemed new. It wasn't until the Software day on Friday that I was in my element. We broke into small groups and designed basic apps for Clotho. My team definitely wrote our program fastest!
6 June-
All of this week was spent working on other HCI lab projects. Michelle and I worked on adding a component to the Green Touch iPhone application. This new component, the Bloom Survey, is meant to take notes on what plants are in bloom at what times. Hopefully, I will be able to use my iPhone programming skills to work on projects with iGEM later in the semester.
13 June-
This week was also spent on another HCI lab project. Michelle and I worked on and application to track the progress of Beetles eating and invasive plant species outside the Wellesley College Science Center. Michelle also started making plans for a "Notebook" design that the BU Wet Lab team can use. Looks like all the iPhone programming will come in handy later this semester!
20 June-
I finally started working on iGEM this week! Consuelo and I started working on BLAST visualizations. Since Boot Camp was three weeks ago, and my bio knowledge is limited, it took a while to get a handle on what we needed to represent. Good thing Kathy (our Wellesley Bio student) could clarify some points for us.
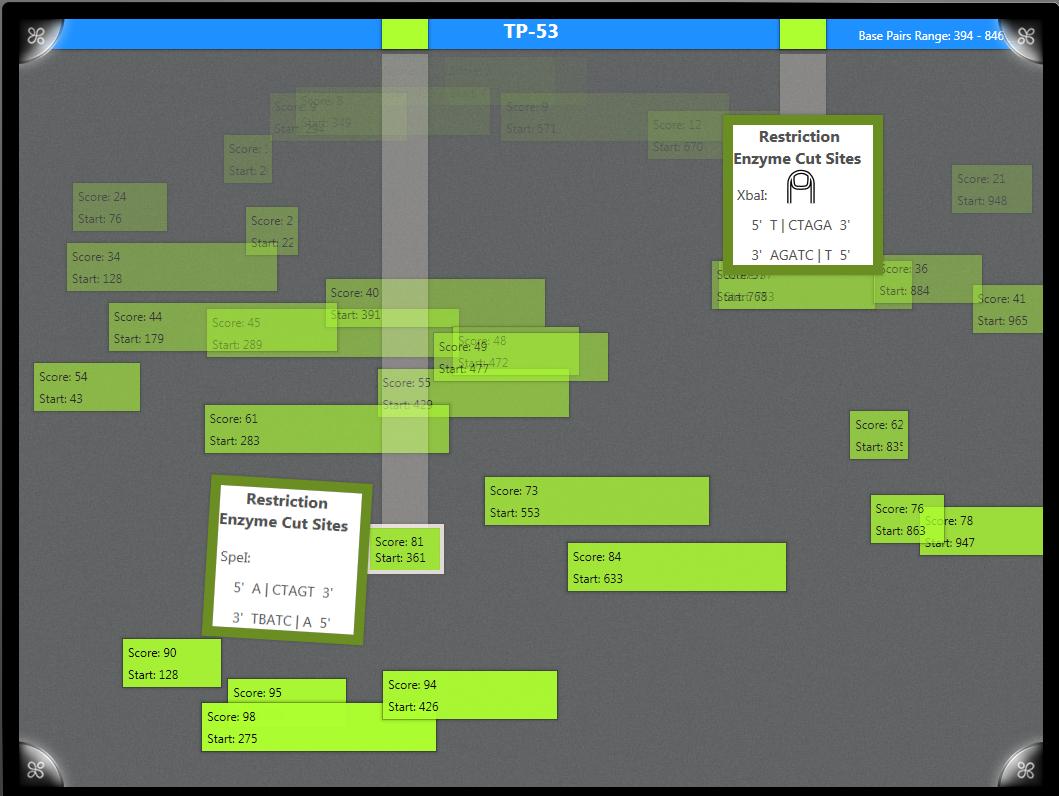
Consuelo and I started on a BLAST visualization meant to display about a hundred results. We used principles of Information Visualization to help users see the score, relative length, and starting point of the various results. Later in the week, Consuelo and I learned that our BLAST visualization did not work very well for Primer BLASTs, which generally have way fewer results, zero being the desired result. So we made a new BLAST visualization for Primer BLASTs. Since we were displaying much less information, screen real estate was not an issue, so we could add more information about each result. We added information about the Restriction Enzyme Cut sites for each result, which will hopefully let the users know how to work with the result better.
See the image to the right for more information.Since the BU Team was coming to Wellesley on Monday, we had to be sure our BLAST visualization was ready to be presented. This required a little bit of work on the weekend, but everything was ready by Monday morning!
27 June-
Monday the BU iGEM group came to see all of our progress on the surface software. We also shared our ideas on an application to help the Wet Lab team in the lab. After sharing these ideas, we had a large brainstorming session with all participants. One group focused on Primer Design, another group focused on overall design, and the last group focused on an application to be used in the Wet Labs. I brainstormed with members in the Wet Lab group. We focused mostly on how users will interact with the application.
We ended the day with a game of Ultimate Frisbee against some other Wellesley College Summer Research Students. See photo to the left.
Later in the week we began focusing on an application, currently titled Lab Buddy. We began researching different styles of interaction techniques. Once we had a stronger grasp on the feasibility of each option we proceeded towards design. We tried to come up with three different designs for each main function in the application. Through process of elimination we decided on a final design and began an electronic prototype.
5 July-
I started today working with Consuelo on the application. But, since the Surface programming required more work I began working again on BLAST. Since we had two visualizations, one for nucleotides and one for primers, it was now time to start working on the back-end. This took a while since Primer BLAST differs from BLAST in many ways. Since no executable could be found for Primer BLAST, we looked at ways BLAST can be used for Primer BLAST. This proved more difficult than originally anticipated since BLAST requires you to BLAST against a database, not a sequence. Megan and I have looked at many work-arounds for this requirement and hope to find something soon.
11 July-
Early this week I found out how to make Primer BLAST work using BLAST through the command line. This step allowed us too begin the visualization for our Primer Designer. We wanted the Primer Designer to be a Microsoft Surface application allowing users to collaboratively design Primers and save them as BioBricks. Ideally, this tool would integrate into Genome Surfer Pro, putting all of our tools into one comprehensive suite. Once we had a design, Casey and I began working on implementing our Primer Designer for the Microsoft Surface.
18 July-
With our Primer Designer front end looking good, Casey and I decided to start working on a Primer BLAST visualization, focusing on multiple genomes. Since out wet lab team works mostly with tuberculosis, we wanted to create a Primer BLAST against five strains of tuberculosis. Since we had the back end working through the command line, we integrated that with our new visualization to have our first fully functional Primer BLAST. However, BLAST is not the only test used when determining the effectiveness of a Primer. So, we talked with Jenhan from the BU software team to take a look at the algorithms he's been working on. Casey and I then took some of his work, and added in much of our own, to add new tests to our Primer Designer, including alignment and hairpinning.
Check out our interns using the Primer Designer on the Surface! See the image on the right.
25 July-
Since our Primer Designer is in such good shape, Orit wanted us to integrate it into Genome Surfer Pro for some upcoming user studies. So Casey and I worked with Mikey on getting our code integrated into the Genome Surfer Pro code. With our code integrate, the HCI Lab began running user studies to see how easily users can interact with the system. After getting some preliminary feedback, Casey and I began implementing changes to our current design in order to make it more user friendly. We plan on having many of the usability issues fixed by Monday so that the BU Wet Lab team can test G-nome Surfer Pro, including our Primer Designer.
1 August-
This is the last week of Wellesley Summer Research so there will be lots of wrapping up to do not just for iGEM but also for the HCI Lab in general. On Monday there was a user test with BU, so much of the week was be spent analyzing those tests and implementing changes by Friday.
We also have a final poster session at Wellesley College to show other research labs what we've been up to. Check out this great poster by Heidi! See photo on the left.
 "
"