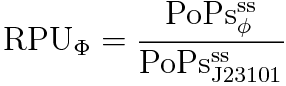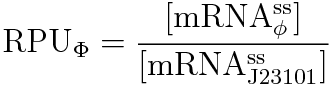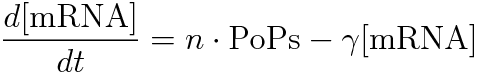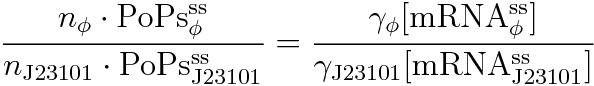Team:Kyoto/Hunger
From 2011.igem.org
Contents |
Project Hunger
Production of needless enzymes is a heavy burden especially when the resource is scarce. This can be reduced by using nitrogen regulatory proteins, NtrB and NtrC† which activate σ54promoter when the supply of nitrogen is not enough. NtrB and NtrC are coded in glnL and glnG, respectively.しかし、今までσ54プロモーターは定量評価されていなかった。 Here, we evaluated the relationships between expression under σ54promoter and that of these genes by the aid of RPU(a relative promoter unit).
† NtrB and NtrC are otherwise called NRII, NRI
ところでこの遺伝子はもともとBioBrickに登録されていたものか、今回新たに作ったもののどちらなのでしょう
http://partsregistry.org/wiki/index.php/Part:BBa_J64978
などがもう登録されているようですが
リンク先を見ましたがどうなんでしょうね とりあえずこの登録されているパーツのシーケンスは見ましたが今回我々が作ろうとしたglnLとは全然違うシーケンスでした 名に由来かは知りませんがE.coliのものじゃないでしょうねby Hashiya
Introduction
For every living thing, needless biological activity is not efficient. Cells must be controlled so that enzymes are produced only when they are necessary. Ammonia is an essential nitrogen source for the bactria. When enteric bacteria are deprived of ammonia, they express glnA to produce glutamine synthetase(GS). Nitrogen is used in the reaction of
Glutamate + NH3 + ADP → Glutamine + ADP + phosphate
GS
The expression of glnA is regulated by several proteins including NtrB, NtrC, Pii.
Fig. NtrCがσ54,RNAポリメラーゼ,DNAの関係図(窒素源あり/なし) Fig. NtrBCの関係図 Fig. Ntr, Pii, GSの関係図
Method
We created two following constructions to measure gene expression depending on the concentration of glutamine. We used BBa_J23101 as a promoter for construction1,which is used as the criteria, and binding sites and σ-54 promoter for construction2, which is object to be measured.
We also measured the amount of mRNA to calculate RPU of the steady state for the each concentration of glutamine. First, we made following preliminary experiment to measure the length of times before steady state.
We cultivated E.coli in M9 media(+ casamino acid) for about 15 hours and dispenced 2.4ml to each tube. Then, we centrifuged these tubes (13,000 rpm , 4℃, 1min) and discarded the supernatant. We added 1.2ml media(- casamino acid) and centrifuged at 4℃ twice. Again, we centrifuged these tubes and discarded the supernatant and added 1.2ml media(-casamino acid) at 37 ℃. We brought up E.coli at 0,5,10,15,20,25,30,60min and extracted RNA and synthesized cDNA. Finally, we used real time PCR.
RPU is defined as follows.
This time, we decided to use the equation (2).
Result
Gragh1 The data, which is not corrected with internal control
- Although the amount of RNA is different first, we think more than the twice difference is not accidental error of the experiment.
- After 15 minutes, expression level greatly changed, but from 0 to 10 min, expression level is steady state.
- We think TBP and PGK are unsuitable for internal control.
- The behaviors of GAPDH and actin is analogous.
Gragh2~5
The data, which is corrected with internal control
- when we applied internal control to GAPDH, the changes of actin are little(×1.0~1.2)
- when we applied internal control to actin, the changes of actin are little(×0.8~1.0)
- We concluded that GAPDH and actin are suitable for internal control.
 "
"