Team:Freiburg/Notebook/6 September
From 2011.igem.org

Contents |
Commons
Miniprep
Investigators: Sandra
Yesterday LB was inoculated with strains (S14, S39-S44 etc.). Miniprep of strains.
Digestion: 2A-assembly
Investigators: Sandra
Digestion of:
- S14 (GFP) pSB1A3
with XbaI and PstI
- S39 (PR1) pSB1C3
- S40 (PR2) pSB1C3
- S41 (PR3) pSB1C3
- S42 (PR4) pSB1C3
- S43 (PR5) pSB1C3
- S44 (PR6) pSB1C3
with SpeI and PstI
incubation for 6 hours at 37°C. Inactivation of enzymes at 80°C for 20 minutes.
Samples were loaded onto a gel and there were no inserts of GFP.
Therefore RFP was digested with SpeI and PstI over night to ligate it with PR1-PR6 tomorrow.
green light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
blue light receptor
Trafo
Investigators: Sandra
Trafo of ligated parts of the 2A-assembly.
Sequence analysis
sequence analysis of our lovtap-Notgate was neagtive:-( So we´ll try the gibson assembly.
PCR
Investigators: Sophie PCR
| Name: Sophie
| Date: 06.09.11 |
| Continue from Experiment (Date)
(Name) | |
| Project Name: Gibson of ♥ and NOT | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | ♥: P97
NOT: P99 |
| 2.5µl | Primer dw | ♥: P98
NOT: P100 |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | NoT, M45 (BBa_Q0400)
original DNA from Edinburgh (BBa_K322999) |
| 0.5 µl | Phusion (add in the end) |
Digestion with Dpn I and purification with PCR purification kit What program do you use?
First 20 cycles touchdown 69°C -0.4/cycle
next 10 cycles touchdown 72°C -0.2 per cycle
PCR products were loaded onto a gel, but there were no bands execpt for not3.
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
Troubleshooting of the modified Lysis genes K124017
Investigators:Theo
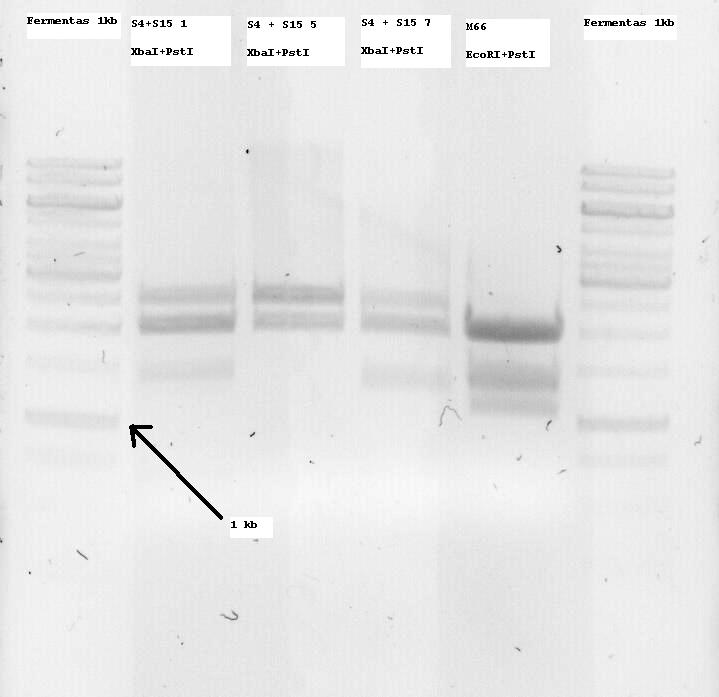
S4+S15 Nr1, 5 and 7 were sent for sequencing. Some problems arose and the sequences seemed to not be correct or only parts of them. So I decided to digest all of the minis and if an insert band (ie about 1300bp, since it is K124017 with RBS) was seen, to extract it from the gel and use it for subsequent ligation with the Promotor.
M66 which is (or SHOULD be -arrghh-) the alreadz modified Lysis genes K124017 was digested with EcoRI and PstI and two bands at around 1100 and 1400bp were seen, so both of them were extracted. I want to clone them on the pSB1C3 Vector and sequence that so that the part is also ready for the parts registry. More on that tommorow.
- Note: S4 is the B0034 RBS (ribosome binding site), S15 is the non-modified K124017 Lysis Cassette without RBS
- Note 2: S4+S15 Nr1, 5 and 7 were digested with XbaI + PstI so that they can be one-step cloned with the temperature sensitive promotor (to be cut with SpeI and PstI and then treated with alcalic phosphatase as well as with the Qiagen PCR purification kit)
INSERT GEL EXTR KIT SOP HERE!!!!
Precipitator
NAME OF YOUR EXPERIMENT
Investigators: NAME
 "
"
 Contact
Contact