Team:Wageningen UR/Safety/One
From 2011.igem.org
(→Safety) |
|||
| Line 32: | Line 32: | ||
{{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | {{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | ||
| - | === | + | === Would the materials used in your project and/or your final product pose risks to the safety and health of team members or others in the lab? === |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | The ''Escherichia coli'' TOP10 strain used in this project is a derivative of the non-pathogenic K-12 laboratory strain. The likelihood of a human becoming ill is therefore low. The most probable route of transmission would occur by accidental aerosol formation or ingestion. The EPA states the strain is badly retained in the human gut, so the chance of it to become pathogenic by mutations is low (EPA, 2011; Jump to References). Mainly for these reasons, this organism is also classified as BSL 1. | ||
| + | When an infection occurs nonetheless, it is important to note that this part of the project involves an oscillation of an extracellular concentration of a quorum sensing molecule, OHHL (an AHL). The OHHL compound is made by the enzyme that derives from Vibrio fischeri’s luxI gene. Quorum sensing molecules play a role in many bacterial infections. This means successful transfer of the oscillator BioBrick device to a pathogen could possibly increase its quorum sensing-related pathogenicity. To get an impression where such pathogenic organisms are found we let the environmental BLAST of NCBI search for luxI homologous regions in Metagenomes (Zhang, Schwartz, Wagner, & Miller, 2000; Altschul, et al., 1997; [[#References|Jump to References]]). In marine environments and in soil of a farm, in the U.S. of America, environmental representative organisms appeared to have a luxI-like gene. By using other BLASTs we found human pathogens with a significantly corresponding luxI gene (Altschul, et al., 1997; Zhang, Schwartz, Wagner, & Miller, 2000; Altschul, et al., 2005; [[#References|Jump to References]]). These pathogens aren’t representatives of the human gut flora, but they might infect humans by means of contaminated water or food. | ||
| + | The pathogens found by the latter Blasts include mainly Vibrio species, for example V. cholerae, capable of causing gastroenteritis. Because of this, the laborants that handle the modified ''E. coli'' will be held on close watch. They should be removed from the lab work in case they have the symptoms of gastroenteritis. Anyone that works with the BioBrick system should have his hands cleaned before eating, as always, to prevent transfer of the system to with Vibrio species contaminated food. Continuing, also two differing fish pathogens harbor and a coral pathogen harbors a gene that significantly looks like the luxI gene of the Oscillatory BioBrick system. | ||
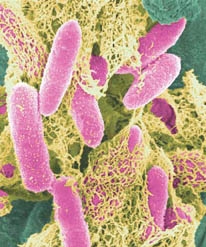
[[File:E-coli-in-color.jpg]] | [[File:E-coli-in-color.jpg]] | ||
| - | '''Fig.2.''' ''A microscopic picture of E. coli cells. The length of a cell is about 2 micrometers. The colours are not real.'' | + | '''Fig.2.''' ''A microscopic picture of ''E. coli'' cells. The length of a cell is about 2 micrometers. The colours are not real.'' |
}} | }} | ||
Revision as of 15:56, 2 September 2011
 "
"