Team:NTNU Trondheim/Alternative systems
From 2011.igem.org
(→Alternative system) |
(→Alternative system) |
||
| Line 1: | Line 1: | ||
| + | |||
| + | |||
==Alternative system== | ==Alternative system== | ||
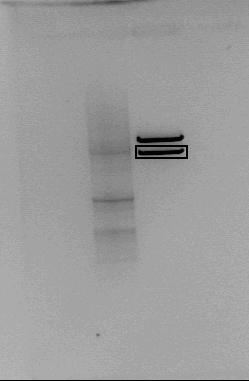
[[File:Puc128_med_LacI-LacP-MC_kuttet_med_EcoRI_090811_exp_wiki.jpg|thumb|Figure 1: Puc128 plasmid with LacI-pLac-mCherry cut with EcoRI. Expected fragments were 2,3 and 3,2 kb. The square indicates the LacI-pLac-mCherry insert that was isolated]] | [[File:Puc128_med_LacI-LacP-MC_kuttet_med_EcoRI_090811_exp_wiki.jpg|thumb|Figure 1: Puc128 plasmid with LacI-pLac-mCherry cut with EcoRI. Expected fragments were 2,3 and 3,2 kb. The square indicates the LacI-pLac-mCherry insert that was isolated]] | ||
Revision as of 11:42, 22 August 2011
Alternative system
From the results, promoter rrnB P1 is not transcribing enough repressor to inhibit pLac-promoter. To test out if the other parts of the system work (LacI-pLac-mCherry), a test system where Pm-promoter controls expression of LacI was made. The wild-type Pm-promoter that was used here is induced by m-toluate.
To be able to transfer the system from pSB1A2 to expression vector pVB20wt they were both cut with XbaI + PstI. The system was inserted into pUC128.
LacI+LacP+MC in plasmid pUC128 was isolated.To retrieve the LacI-pLac-mCherry insert pUC128 was cut with EcoRI and the fragments were separated on gel. The insert fragment were cut out and extracted from the gel as shown in Figure 1.
To transfer it to pVB20wt, pVB20wt was opened using EcoRI and then treated with CIP to avoid religation, and was then ligated to the EcoRI-digested insert fragment.
 "
"