Team:Michigan/Notebook
From 2011.igem.org
(→8/16/2011) |
(→8/09/2011) |
||
| Line 969: | Line 969: | ||
==8/09/2011== | ==8/09/2011== | ||
| + | |||
| + | ''Alena'' | ||
| + | |||
| + | ⇒ Miniprep 2 mL of both His-tagged ZFPs (His-Gli1-nCFP: BBa_K323058 and His-Zif268-cCFP: BBa_K323069) from 8/04/11 | ||
| + | |||
| + | ∴ for BL21 transformation | ||
''Candy'' | ''Candy'' | ||
Revision as of 22:13, 17 August 2011

Calendar of Events
Protocols
Notebook
5/13/2011
Kevin, Ben, Candy, Brian, Alison
⇒ Unpacked supplies from last year
⇒ Restocked lab and created inventory
⇒ Training protocols written
5/19/2011
Kevin, Alena, Ben, Namun
⇒ PCR machine is now operable. Basic programs have been written, but not all the functions of the machine are known. No protocol for using the PCR machine has been written.
⇒ Usage of the autoclave was determined. No protocol exists for autoclaving yet.
⇒ 25 plates were made using 10g LB agar stock, 250mL deionized water, and 250μL ampicillin from vials left over in the stupid white box in the freezer. Original protocol called for distilled water-abbreviation "dH2O" was misinterpreted to mean deionized. Not known if the plates are viable due to this error. House-made protocols will use the following abbreviations to avoid this confusion:
∴Distilled water: dH2O
∴Double distilled water (from the machine): ddH2O
∴Deionized water: DI water or diH2O
A protocol has been written for plate production. As it depends on the autoclave protocol, however, it cannot be considered complete.
⇒ Simple inventorying took place, with the locations of pipettes, ethanol, hardware, and the master gas shutoff(!) determined.
⇒ Inventorying will continue tomorrow, and a PCR reaction may be attempted. Protocols for the PCR purification kit (Miniprep) will be modified during this time, assuming the PCR reaction proceeds. No method currently exists to determine PCR outcome, as no gels or gel mix is available in the lab. Along these lines, it may be beneficial to find an ultraviolet spectrophotometer to run these tests.
5/20/2011
Amy, Ben, Candy
⇒ Cells were transformed with extra 173 pGLO vector in accordance with the QT protocol. This used stock competent cells found in the refrigerator. Protocol modifications will be made accordingly.
⇒ Access to room 3152 is crucial, and no means of entry currently exists. This will be imperative if we require ddH2O, an autoclave, LB stock, a normal scale, large glassware, or any number of other items.
⇒ Due to lack of access to 3152, protocols requiring sterile water cannot be tested. Sterile water exists in 3151A, but is 3 years past its expiration date.
⇒ No source of agarose or ethidium bromide has been found, so electrophoresis protocols cannot be tested. Interestingly, there exists plenty of loading dye.
⇒ Large amounts of stock compounds IE ethanol belong to the room and are not owned by us. Currently they are being used. This may cause problems, especially if we start using more expensive materials we still don't own.
On a side note, the contrast between the physical sign-in sheet in 3151 and the online form on ctools is rather confusing-it may be better to just use the online one, and have people log their hours individually.
5/22/2011
Ben ⇒ Two containers of 500mL LB broth were produced and autoclaved. 12.5g of LB powder was combined with 500mL water for each container.
5/24/2011
Ben ⇒ Six Erlenmeyer flasks of diH2O were autoclaved and are now sterile. Both sets of glassware reside on the shelf next to the incubator.
5/25/2011
Kevin, Alison, Ben, Chris
Had discussion with Dennis and Mark from the USB labs, learned some important points.
⇒ We aren't allowed to just "use" chemicals that are already in the USB that don't belong to us. I am working on ordering more of the basics right now. We are allowed to use the equipment in the room. However, if you don't know how to work a piece of equipment, please ask first!
⇒ Dennis and Mark are two very knowledgable supervisors who are conveniently generally very close to our lab room. If there is any equipment that you need assistance with, you can try to ask them. They are in an office close to our lab, I believe in room 3159. Just knock and be polite!
⇒ There are been some theft issues in the USB lately, so we need to be careful with keeping our door locked. Also, Dennis and Mark don't necessarily know our faces, so they won't necessarily know that we aren't actually trying to rob the USB. Additionally, our own Marc wants to know when we will be in the lab so if possible he could come in and help. If you are going to be in the lab unplanned, please email both Marc and CC Dennis.
⇒ A few more safety things when it comes to the lab- the outside door locks at night (I believe after 9). Make sure you bring your Mcard so you can get into the building after hours if that is needed. Also, the elevators in the USB change at night. One goes from P up to 4 and the other goes from P down to parking. Just keep this in mind. If you have your Mcard, you'll be able to get around the building.
⇒ A few other people that won't be expecting us if we are in the lab all night are the custodial staff, who will most likely call DPS on us. While this would make for a fantastic story, we don't want it happen. If there comes a time when we are in the lab past midnight, also let Dennis know at a decent hour so he can ensure we don't get DPS called on us.
Kevin and Alison tried to run a gel today, only to find that we lacked the necessary reagents and some equipment. We were able to borrow some equipment from Mark: microwave, transilluminator, and an autopipetter. Alison will buy some reagents tomorrow and we will try again.
5/26/2011
Kevin, Alison, Ben, Namun, Candy, David
⇒ Gel materials have been purchased by Alison the Great, including:
∴Agarose ∴Ethidium Bromide ∴TBE Buffer
⇒ A stock of 1% by concentration TBE buffer was generated. This will be used for all standard gels. Sterility with it is recommended, but not completely necessary.
⇒ Courtesy of Kevin, an instructional session on gel production was completed. A rough protocol was written, to which a refined version should be finished by the end of Friday 5.27.11. More practice with these methods would be of benefit to everyone intending on participating in lab work, however, so more sessions should be planned.
⇒ The transilluminator was tested. It works. Anyone running a gel should take a picture of it and upload it to the wiki.
⇒ The shaker/incubator/water bath was also tested. It's not clear if it genuinely requires water to exist in the bottom or if it can simply function as an incubator; when being set at 37°C, however, it seems to randomly rise above 38°C, then emit irritating beeping noises. Exactly how this device operates is still under investigation.
5/31/2011
Ben
Forever alone :'(
⇒ Gel protocol has been tested and a revised version will be uploaded.
⇒ 6 microliters of DNA ladder was used without dilution; this may be a bad idea. Please note the juice+ladder is in a clear centrifuge tube.
⇒ Protocol for gel viewer was tested. Please note the UV source itself is the true "transilluminator"; the entire machine should hence called the gel viewer.
6/1/2011
Ben
⇒ Gel protocol has been tested a second time without success. It appears mixing is absolutely crucial to all reagents, including the DNA; this was not stated in the protocol. No protocol currently exists for making 1% TBE buffer, which we are about to run out of; assistance with this would be appreciated.
⇒ Work has started on a do-it-yourself cold block for storing enzymes made out of solid ice. Current results look bad.
⇒ Preliminary designs for a transport vector that can carry surface display components is underway and is currently in the "argument" stage.
6/5/2011
Ben
⇒ Gel protocol has been tested a fourth time with success. Phusion from the freezerwas used in a rather haphazard way to generate PCR product, and a distinct band was observed. It should be noted that he TAQ solution is now dead for training purposes, but its buffer is receptive to other polymerases. This is completely useless information.
⇒ Enzymes from the Lin lab are now consolidated in the USB. We now also have a nice set of pipettes *borrowed* from the Lab.
⇒ Unbeknown to most, electric autopipetters exist which are portable and easy to use. They are in the cabinet. The current "plug in" compressor-based one is currently out of order as the filter is clogged; this is being investigated further.
6/8/2011
Chris, Narun, Ben
⇒ A digestion test was attempted on remains of the experiment from three days ago. This was completed as a training exercise. Results are still inconclusive. Leftover GFP was cut with XbaI and run on a gel next to a control of uncut GFP "just to see what would happen."
6/10/2011
Josh, Ben S, Chris, David, Ben P
⇒ Cryostocks for 10 different strains [2 Original INP Samples (non-standardized), GFP, linker(-80 and -20 samples), GFP-OmpA ligation (Resistance:K), Omp A, and a constituitive promoter] were taken out and grown overnight in the shaker for 16 hours. Stocks of old competent cells in the -80 were thawed, transformed with promoter J23199 from the 2009 iGEM distribution, plated, and grown overnight.
6/11/2011
Chris, Josh
⇒ All of the strains grown from the frozen stocks were cloudy. Control tube was clear. So, frozen stocks appear to be in good shape.
⇒ The plate for the transformed old competent cells did not appear to have any growth. Let incubate for another day.
⇒ A miniprep was performed on the ten strains grown from frozen stocks. 500 microliters of the original growth was stored in the fridge while 1.5 ml from each was used in the miniprep. The resultant DNA from the miniprep was stored in the freezer overnight.
6/12/2011
Alison, Ben P, Kevin
Note that Ben P is not alone for this post and no sad smiley faces should ensue.
⇒ Some basic lab prep was accomplished tonight
⇒ More 1xTBE solution was made (simple dilution of the 10x TBE with diionized water)
⇒ LB + AMP plates were made, along with LB + KAN plates
⇒ 2 400 mL LB solutions were made using 400 mL of diionized water and 18 g of Agar. These were autoclaved on the 20 minute liquid cycle. After they had cooled, 400 microL of AMP was added to one solution and 400 microL of KAN was added to the other solution. Once the bottles were cool enough to hold, the solutions were poured into Petri dishes to make plates. Sterile technique was used. Plates were allowed to rest and solidify overnight and were labelled and placed in the refrigerator to store. LB + AMP plates were labelled with blue marker and LB + KAN plates were labelled with red according to standards found online.
6/15/2011
Chris, Ben P, Alison
⇒ The QT comp cell production protocol was tested. 7 aliquiots of competent cells were produced (it was supposed to be five), which is rather unusual and will have to be investigated. Innoculation occured on the 14th at around 2200, leaving the growth time at around 17 hours. It should be noted the spectrophotometer is rather quirky and may require a larger amount of liquid than originally thought; this will be investigated as well, and the effected protocols modified.
⇒ 0.1M calcium chloride stock was produced a 1M stock-we don't have any solid CaCl2 laying around, so when we run out of the parent 1M stock we're done.
6/18/2011
Chris, Josh Inoculated 3 different RBS strains from cryostock. This is just something to do until we have the competent cells and spectrophotometer working.
6/19/2011
Ben, Alison
⇒ Comp cells were tested a third time; stocks thawed from the -80C freezer were transformed along side other stock from the fridge. 100 microliters of each were transformed with leftover pGLO, heat shocked in accordance with the respective protocol, and incubated for ~60 minutes in 1mL LB. The stock from the fridge appears to be dead, as after the heat shock protocol it refused to grow in the LB; the -80C stock made the solution cloudy. Both were plated onto AMP plates.
⇒ Sunday night lab prep consisted of mostly just cleaning. The liquid waste bottle was autoclaved, as were some other dirty glassware. Autoclave behaved very nicely. General tidying up of the lab was done and hopefully the rest of the time finds it satisfactory. It didn't appear that any stock solution or plates needed to be made. I will be out of town for next 2 Sundays, will maybe pass the baton to LJ for Sunday lab prep? Should probably actually talk to him.
6/22/2011
Marc, LJ, Chris, Alison
Marc shared with us his basic protocol for making competent cells; the protocol is included also.
⇒ Day One: Start culture of cells (approximately 5 mLs?)
⇒ Day Two: Subculture Culture: Dilute it to 1/40 concentration = Grow in incubator for 1-1.5 hours Option #2: Dilute it to 1/100 concentration = Grow in incubator for 1.5-2 hours
⇒ Let grow until OD(540)= 0.4 (approximately)
⇒ Don't overgrow cells! Culture medium should be just visibly cloudy (when swirling the container, should be able to see swirls of cells)
⇒ Spin cells down in centrifuge, wash pellet with 0.1 M MgCl2
⇒ Spin cells down again, wash/resuspend pellet in 0.1 M CaCl2
⇒ The final volume should be about 1/5 that of the subculture volume.
Note: need 200 microL per transformation
Detailed Protocol:
⇒ Day 1: Chip off some DH5alpha from the cryostock and put it into fresh media. Place in incubator at 37 degrees (C) overnight.
⇒ Day 2: Media from last night was definitely cloudy. 200 mL of the incubated media was split into 2 100 mL amounts in two separate flasks. 2.5 mL of incubated media/cells were added to each new flask. This was placed back into the incubator for another hour. After only an hour, the media/cells were at the right cloudiness. We didn't bother actually checking their concentration. The media/cells were transferred to 4 50 mL centrifuge tubes and put in the centrifuge at 5000 RPM for 5 minutes at 4 degrees (C). After 5 minutes in the centrifuge, the media was poured out (a cell "pellet" was left at the bottom) and the four tubes were placed upside down on a clean Kimwipe to ensure all the media drained out. About 30 mL of 0.1M MgCl2 was added to one flask; that flask was shaken until the cells were resuspended, and then that flask was emptied into the next one and shaken to resuspend the next cell pellet. This continued until all the cell pellets were dissolved in one flask. This was placed back in the centrifuge (with one flask of water to balance) on the same setting as before. After 5 minutes, the MgCl2 was drained out of the flask and about 40 mL (about 1/5 of the original volume from Day 1) of 0.1M CaCl2 was added to the flask. This was mixed, labelled, and placed in the refrigerator.
Other notes from the day:
⇒ To Do: Make and sterilize 50% glycerol containers to store cells in at a future time. After the cells are added to the glycerol, you want the entire solution to be at 15% or so (why making 50% tubes would be a good idea). This would be good for making additional stocks of DH5alpha.
⇒ Words of Wisdom from Ammerlaan: Biology is a "meatball science"... Not everything has to be completely exact! As long as it is about right, it should turn out correctly. Also, sterile technique is important, but in some instances it shouldn't be completely stressed over. When making cells, sterile technique is very crucial at the beginning when you have very few cells grown. Contaminating a small amount of cells could be a large problem, whereas contaminating a huge batch of cells might not affect them much. If you get a chance to see the Ammerlaan version of sterile technique, I recommend it. Also, if you are ever pouring anything in his presence, please be careful. This is one of the things he is particular about!
⇒ Autoclave stuff: Should start an autoclave bag for all the small things to be autoclaved as waste (such as tips, tubes, etc.). Once we fill this bag up to 70%, then we can autoclave it as waste. Bags that have been autoclaved can be thrown away in the large trash can by the autoclave. This has been verified by Mark.
⇒ Using the spectrophotometer: Entire samples of DNA miniprep were mostly < 50 microliters, so the entire sample was used, in addition to around 20 microliters of water to measure DNA concentrations. Then, the contents were transferred back to 1.5ml tubes for storage and labled DNA concentrations.
6/23/2011
Chris, Ben S
⇒ We first measured DNA concentrations, in micrograms/microliter: INP 1.1: 0.006 INP 1.2: 0.013 OmpA: 0.011 GFP: 0.005 Promoter: 0.017
⇒ A digestion was run for OmpA and GFP. OmpA was digested with E and S, and GFP was digested with E and X.
⇒ A Gel was performed on the digestion. The results did not look as expected: -Gel Pic Here-
6/24/2011
Kevin, Candy, Alena, Chris
⇒ ddH2O was "liberated" from a source on campus. It's in the big barrel by the windows in the lab.
Chris, Ben P, Ben S
⇒ Digestions were attempted again with the following samples from cryostock:
OmpA-GFP 1
OmpA-GFP 2 (1 and 2 from last year miniprep)
OmpA-GFP (6/11/11)
INP-Linker-GFP
INPNC 1
INPNC 2
Linker
Promoter
Candy, Alena
⇒ Zinc finger protein parts BBa_K165006 (ZiF268-HIV) and BBa_K165007 (Gli-1) with ampicillin resistance arrived from registry. (Request sent on June 5, parts shipped on June 21.)
Plate each on 3 LB/Amp plates, incubate 37 degC overnight.
6/25/2011
Ben
⇒ The follow overnights were prepared with 5mL LB and 5μL ampicillin:
AIDA-1
GFP
OmpA
Promoter
RBS
ZnF Gli-1 (BBa_K165007)
ZnF Zif268-HIV BBa_K165006)
⇒ A 50mL stock of LB was created out of the larger stocks; please use this smaller stock instead. A stock of 50% glycerol was also made and resides in the refrigerator.
⇒ Please wash out the photocuvettes with ddH2O between uses!
⇒ When using ampicillin or other antibiotics, please keep them COLD. They will degrade otherwise.
6/26/2011
Ben P, Chris, Alena
⇒ Miniprep (1 of 2) Zif268-HIV and Gli-1 from overnight on 6/25/2011.
Concentrations:
∴Dilution: 5uL samples:55uL diluent
∴Gli-1: 0.049 ug/uL ∴Zif268-HIV: 0.080 ug/uL
Placed in -20degC freezer.
6/29/2011
Candy, Alena
Miniprep (2 of 2) of Gli-1 and Zif268-HIV from overnight on 6/25/2011. The two minipreps (today and 6/26/2011) are combined for digestion.
Digest minipreps of the following with EcoRI and PstI:
Gli-1
Zif268-HIV
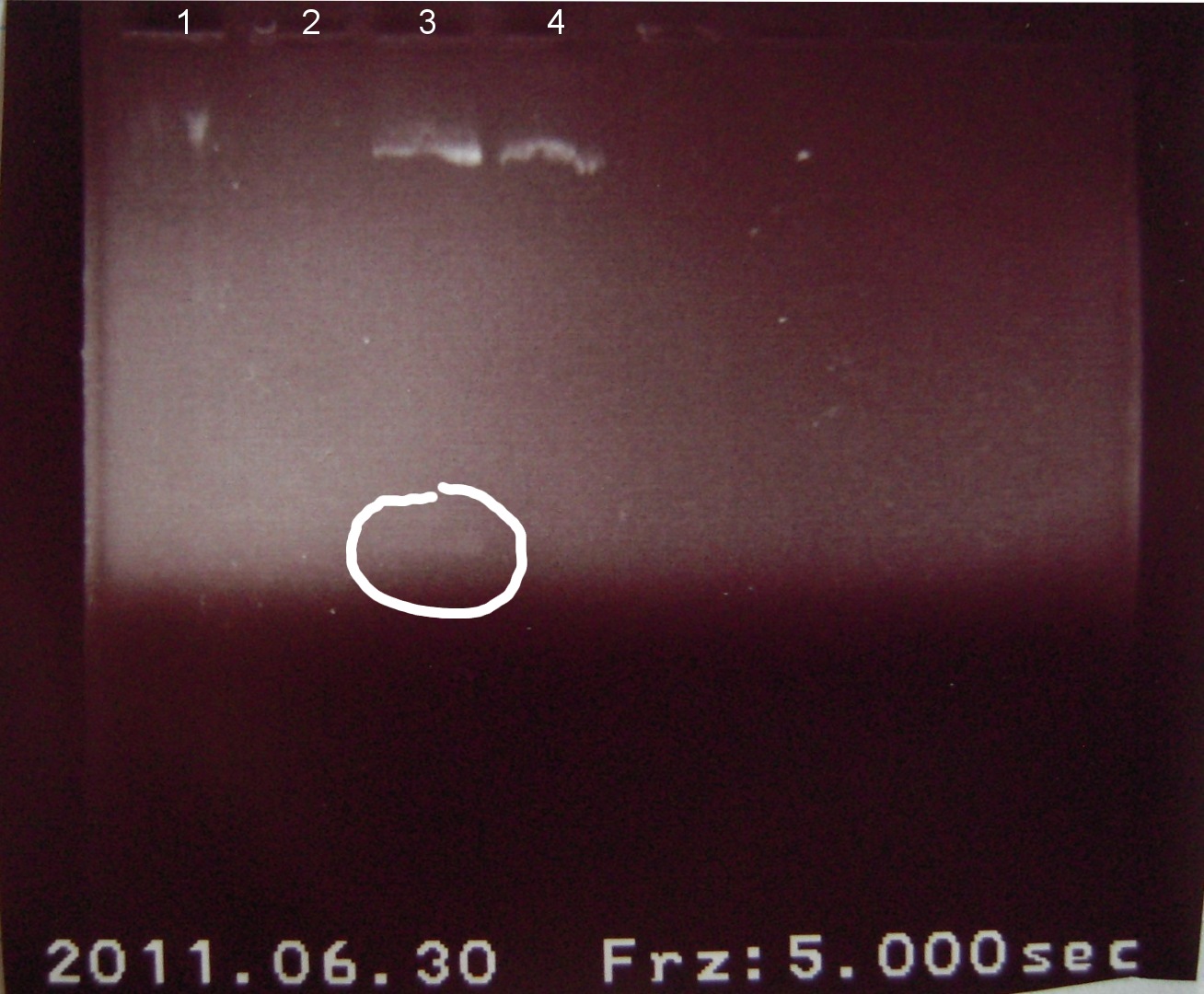
6/30/2011
Alena, Candy
Gel electrophoresis of digests on 6/29/2011
Lanes:
1. Ladder 1kb
2. "Empty" Digestion: solution incubated without DNA
3. Zif268-HIV digestion solution (293 bp)
4. Gli-1 digestion solution (563 bp)
∴Ladder cannot be resolved (perhaps no loading buffer added).
∴Band on lane 3 may be fragment of interest for Zif268.
∴No fragment apparent for Gli-1.
Josh
Digests: INP+linker 1.1, INP+linker 1.2, INP + linker + GFP, linker + GFP 1 (4.5 ul needed), and linker + GFP 2.
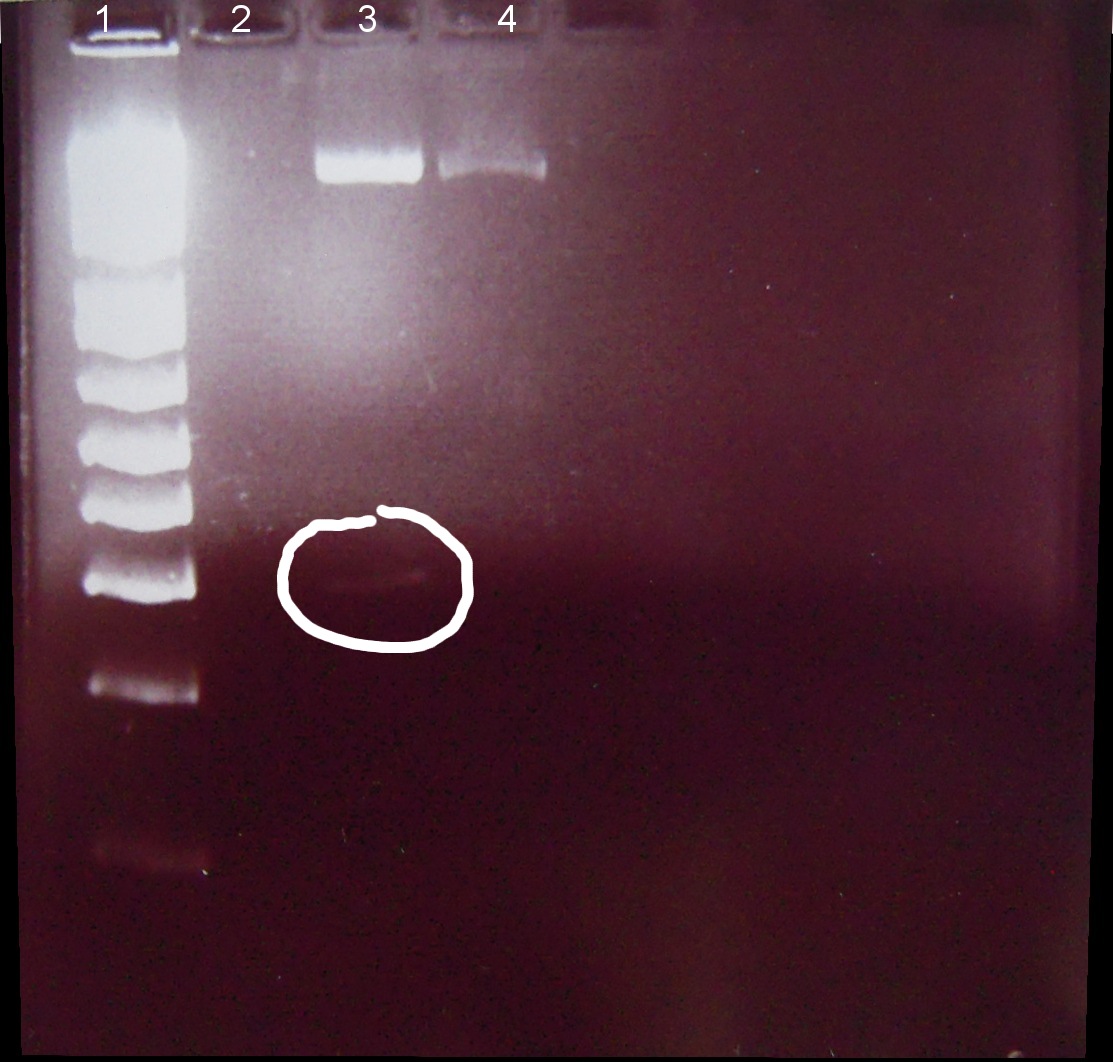
7/01/2011
Alena, Candy
⇒ Loading dye and digestion mixture amount doubled in attempt to increase band strength.
1. Ladder 1kb
2. "Empty" digestion
3. Zif268-HIV digestion solution (293 bp)
4. Gli-1 digestion solution (563 bp)
∴Zif268-HIV band of ~300bp visible.
∴No band of ~550bp apparent for Gli-1, possible reasons: 1. ethidium bromide haze obscured very weak band, 2. failed digestion. Will repeat miniprep and digestion of Gli-1 at later date.
⇒ Inoculate Zif268-HIV and Gli-1 cultures and incubate on shaker overnight in LB/Amp in preparation for -80degC storage.
7/02/2011
Alena, Candy
⇒ Miniprep Gli-1 (follow up of 7/01/2011) for later digestion and gel electrophoresis.
EB elute used 30uL instead of 50 uL in an attempt to increase plasmid concentration.
⇒ Store Zif268-HIV (3 aliquots of 1 mL each) and Gli-1 (2 aliquots of 1mL each) in -80degC (follow up of 7/01/2011).
7/03/2011
Ben P.
⇒ Two parts were extracted from the wells and 2μL transformed using the heat shock protocol.
-pSB1K3, plate 1, well 5A (we already have stocks of this as well)
-BBa_J04500, plate 4, well 12A
The latter part contains an IPTG inducible promoter and RBS fused together. The default insert for pSB1K3 contains RFP bound to the same promoter/RBS. It is not known if RFP will be expressed properly, as the promoter depends on both lacI and CAP, which may or may not be present in the DH5α strain. Experimentation with these two parts will continue through the next week, with the primary objective to insert this IPTG pro/RBS system into AMP,KAN, and maybe other plasmids for use by all subteams for expression purposes.
⇒ IPTG stocks exist, but must be diluted down.
⇒ pSB1K3 is contained in a stock from last year. So now we will have two stocks.
⇒ BioBrick primers must be ordered. Still. Obtainment of these primers will permit the expansion of protocol and method flexibility.
7/04/2011
Ben P.
⇒ Overnights of the two parts were purified today. 4 stocks exist total, two of each part. Concentration of one stock was measured to be 47ng/μL, which is rather disappointing.
⇒ No concentration measurements were taken on the other three stocks. Please refrain from measuring stock concentration with existing (used?) cuvettes, as the cuvettes are not designed for multiple uses and contamination could result.
7/07/2011
Ben S.
⇒ Transformed 15 parts from distribution: 5 promoters (constitutive), 3 QPI's, 2 Carrier Proteins, 2 Arac/Pbad promoters, 1 lacPO+RBS intermediate, 1 plasmid. These parts will be used to construct a variety of expression systems for our surface display protein.
Transformation Protocol:
1) Chill 200 uL cells and 1 uL DNA ice 20 m
2) Heat shock 1 m @ 43 deg C
3) Plate Cells or Recovery Followed by plating
⇒ Transformed cells were plated in two sets: one that was allowed a recovery period in LB broth of 30 minutes and one directly plated after heatshock for the purpose of optimizing our transformation protocol.
Josh
Digests continued.
7/8/2011
Ben S.
⇒ The transformations from 7/7 were not very successful due to drying caused by the incubator fan. Repeated transformations and plated only directly (no recovery).
7/9/2011
Ben S.
⇒ Colonies transfered to LB broth for growth. QPI, an AraC/pBAD, and our OmpA protein did not grow, likely due to the lack of a recovery period before plating with Kanamycin.
Josh
Gel 1
1 - ladder
2 - INP + Linker 1 EcoR1 + Xba1
3 - INP + Linker 1 Xba1 + Pst1
4 - INP + Linker 2 EcoR1 + Spe1
5 - INP + Linker 2 Spe1 + Pst1
6 - INP + Linker + GFP EcoR1 + Xba1
7 - INP + Linker + GFP Xba1 + Pst1
8 - old ladder
Gel 2
1- ladder
2 - Linker + GFP 1 EcoR1 + Xba1
3 - Linker + GFP 1 Xba1 + Pst1
4 - Linker + GFP 2 EcoR1 + Xba1
5 - Linker + GFP 2 Xba1 + Pst1
7 - old ladder
7/10/2011
Ben S.
⇒ Transformation of the failed transformants (7/8) repeated. Miniprep was carried out on the successful transformants and stocks of the DNA both placed in the fridge and frozen in glycerol as stocks.
Josh
Conclusions from last gel:
INP + Linker (K157013)
• 920 + 40 = 960 bp (2428 bp)
• redo digests
INP + linker + GFP (E0040)
• 920 + 40 +720 = 1680 bp (2079 or 2428 bp)
• good to go
Linker + GFP
• 40 + 720 = 760 bp (2079 or 2428 bp)
• don't know, don't care
Next digests: INP+linker 1.1, INP+linker 1.2, constitutive promoter 1, constitutive promoter 2, RBS, RBS 1, RBS 2, and RBS 3
7/13/2011
Alison, Kevin, LJ, and Brian
The DNA Printing team came into the lab to start testing the plain slides from Mycroarray to see if E Coli cells would naturally stick to the slides and what buffers should be used.
⇒ Looked up a PBS recipe for our first buffer. We added 2.62 g NaH2PO4(H2O), 11.5 g Na2HPO4, and 43.84 g NaCl to 450 mL dH2O. The pH was adjusted to 7.4 using 4 M NaOH (odd because the protocol only told us to adjust it using HCl). The final volume was brought to 500 mL. We then made 400 mL of 1xPBS buffer by diluting it with dH2O.
⇒ DH5alpha cells were centrifuged and the liquid drained from it. A small amount of the dense cells were placed in a 50 mL centrifuge tube and nearly 50 mL of 1xPBS media was added to the tube.
⇒ One blank slide was placed in the centrifuge tube with cells and PBS and let sit for 20 minutes.
⇒ After 20 minutes, we removed the slide without rinsing and checked it under the microscope. There were definitely E Coli cells stuck to the slide. This is most likely due to us choosing not to rinse the slides.
7/14/2011
Josh and Amy
Digests and Gels
1 - old ladder
2 - INP + Linker 1 EcoR1 + Xba1
3 - INP + Linker 1 Xba1 + Pst1
4 - INP + Linker 1 EcoR1 + Spe1
5 - INP + Linker 1 Spe1 + Pst1
6 - INP + Linker 2 Xba1 + Pst1
7 - INP + Linker 2 EcoR1 + Spe1
8 - RBS EcoR1 + Xba1
9 - RBS 1 EcoR1 + Xba1
10 - RBS Spe1 + Pst 1
11 - RBS 1 Spe1 + Pst1
12 - new ladder
1 - old ladder
2 - RBS 2 EcoR1 + Xba1
3 - RBS 2 Spe1 + Pst1
4 - RBS 3 EcoR1 + Xba1
5 - RBS 3 Spe1 + Pst1
6 - constitutive promoter 1 EcoR1 + Xba1
7 - constitutive promoter 1 Spe1 + Pst1
8 - constitutive promoter 2 EcoR1 + Xba1
9 - constitutive promoter 2 Spe1 + Pst1
10 - new ladder
7/15/2011
Ben
⇒ Both stocks containing ~20µL of 04450 and 04500 were completely digested with 1µL E and P for two hours at 37°C. A total solution volume of 30µL will be produced.
7/16/2011
Ben
⇒ Both digests containing ~20µL of 04450 and 04500 were run on a gel. The gel looked abysmal. A second gel was run. It also looked abysmal. No bands corresponded to any correct base pair amount. A second purification and digestion must be attempted.
⇒ In response to this, a second overnight was prepared in order to attempt the digest again. The next digest will use an NEB protocol.
7/17/2011
Ben
⇒ The second digest was purified and digested with E and P using the following solution:
32.5µL diH2O
10 µL DNA
5 µL NEB3
0.5 µL 100x BSA
1 µL EcoRI
1 µL PstI
2 hour digest at 37°C, with a heat inactivation of 15 minutes at 80°C.
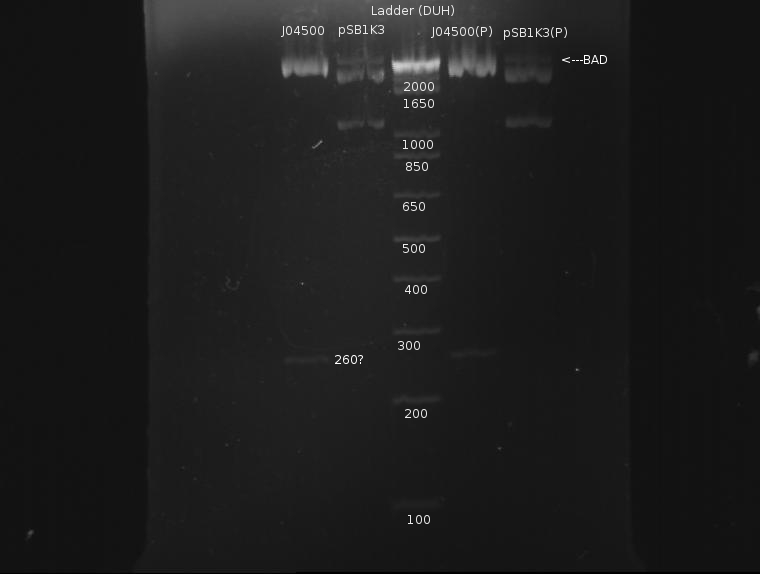
The gel looked like this:
Apart from a slight band around pSB1K3 indicating incomplete digestion, the digestion was extremely clean. This protocol was recommended by both the Knight laboratory and NEB, and as such it seems to work well.
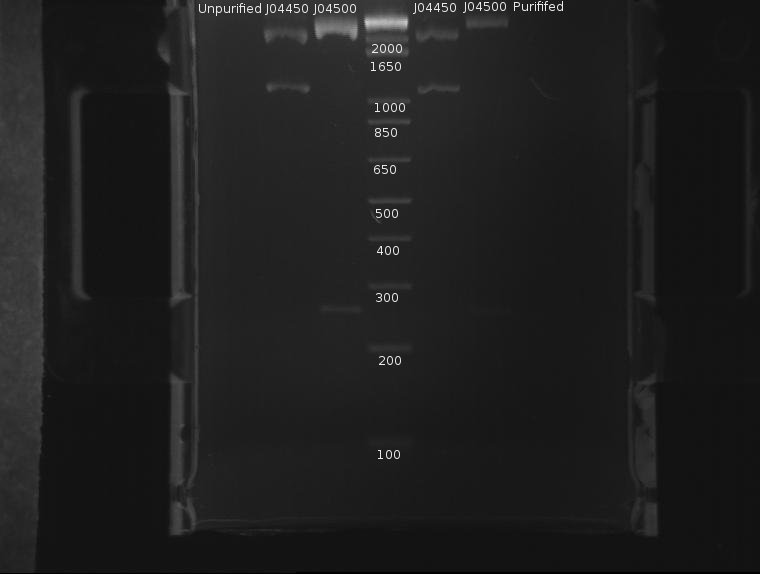
⇒ The DNA was then purified using a Quigen PCR purification kit. DNA was eluded in 30µL EB. A second gel was run with 8µL pre-purification and 8µL post-purification DNA for two reasons:
∴J04500 appears too long: it should be 220bp.
∴The purification efficiency of the PCR kit is currently unknown when purifying digests, and this must be tested-especially since a J04500, a 220bp segment, is pushing the 200bp limit of the kit.
∴Increased resolution of the undigested component of pSB1K3 would be useful.
J04500 and J04450 refer to the inserts, not the part itself. The important component of J04450 is the plasmid, pSB1K3. This is a typo on the drawing. While the bands representing J04500's plasmid pSB1K3 (~2200bp) do possess an uncut component, it appears very small. The worrying line corresponds to the insert around 200-300bp (J04500.) While this is likely the correct length when the prefixes and suffixes for assembly sandards are taken into account, the band itself is very faint; obviously, a large amount was lost in the purification process. As only 20µL or so of this sample remain, it remains to be seen if this amount is enough for use in a ligation.
Two significant amendments were made to the gel protocol this time:
∴As the DNA ladder solution is EXTREMELY concentrated, which is part of the reason for the terrible, splotchy bands in the previous gel. It was diluted down 20-fold to a concentration of 50ng/µL. This stock is very functional and resides in the freezer.
∴An 8-tooth comb was used, as it seems to produce more precise and easy to measure bands. This should be used as standard unless many samples are used.
Josh and Amy
PCR of INP + linker 2 performed using last years screening protocol.
7/18/2011
Ben
⇒ Calculations indicate insert concentration to be to low. A second digestion was attempted, this time with more volume to hopefully give a higher yield:
65 µL diH2O
20 µL DNA
10 µL NEB3
1 µL 100x BSA
2 µL EcoRI
2 µL PstI
__________________
100 µL total reaction
2.5 hour digest at 37°C, with a heat inactivation of 15 minutes at 80°C.
⇒ Purification of ~90µL of this solution was eluded in ~50µL buffer EB. A gel was run pre- and post-purification:
Again, the plasmid appears to not have cut completely; otherwise, the results look fairly nice. Purified DNA appears to have roughly the same concentration as the unpurified DNA, which is rather dissapointing, as it indicates almost half the DNA was lost in the column. Still, ~40µL remain, and I will attempt a ligation.
⇒ Ligation proceeds as follows. The remaining 3µL is taken up by 2µL T4 ligase buffer and 1µL T4 Ligase:
∴L1(1:1), 5µL pSB1K3, 5µL J04500, 7µL H2O
∴L2(3:1), 3µL pSB1K3, 9µL J04500, 5µL H2O
∴L3(6:1), 2µL pSB1K3, 12µL J04500, 3µL H2O
∴L4(8:1), 1µL pSB1K3, 8µL J04500, 8µL H2O
Unfortunately, a possibility exists that the concentration of used DNA will be too high, as NEB recommends keeping concentrations at 1-10ng/µL; DNA from the purification is estimated at 30-50ng/µL. It remains to be seen if this will have an effect, but seeing as how the stocks were used almost entirely for this procedure, this may be unfortunate. Dilution of the remaining stocks may be necessary if this is to be attempted again, both to increase efficiency and stretch the remaining materials.
The T4 ligase buffer is old, but expires in 2013. The amount of ATP left in the buffer is unknown, since to my knowledge no reactions with this buffer have been attempted since September or October '10. Transformation will be attempted with Heat Shock 2.00 protocol.
⇒ J04500 and pSB1K3 were streaked out on new KAN plates to produce new colonies. They currently reside in the incubator, top shelf on the left.
7/19/2011
Ben
⇒ J04500 and pSB1K3 grew into nice plates of mixed colonies (red, white) indicating both inserts made it into plasmids. To screen against the insert, white colonies containing J04500 were picked and plated in arrays based on ligation number onto two plates-one AMP, one KAN. Colonies containing pSB1K3 will die on AMP, but live on KAN.
7/21/2011
Josh
INP + Linker 2 PCR purified
Conclusions from last digest:
• something is wrong with INP, the Spe1+Pst1 digests are producing 2 bands
• need to digest all INP constructs
Next Digests:
• 4 digests each of INP miniprep 1, INP miniprep 2, INP PCR 1, and INP PCR 2
• 4 digests each of linker miniprep 1 and 2
7/23/2011
Ben
⇒ Arrays of Ligations L1, L2, L3, and L4 were compared between AMP and KAN plates to see which colonies survived. A total of 10 colonies were found surviving on KAN, but not AMP.
⇒ Colonies were scraped and re-suspended in 50µL sterile water. A PCR mix containing Phusion enzyme, dNTPs, Buffer, VF2/VR primers and diH2O was produced and 16µL added to 11 PCR tubes. 2µL suspended colony was added to the 11. The cells were placed in the PCR machine and run with the following program:
∴15 min 98°C (break open cells-initial)
∴30 sec 98°C (melting-start of cycle)
∴30 min 57°C (annealing)
∴1 min 72°C (elongation-end of cycle)
∴Cycles: 39
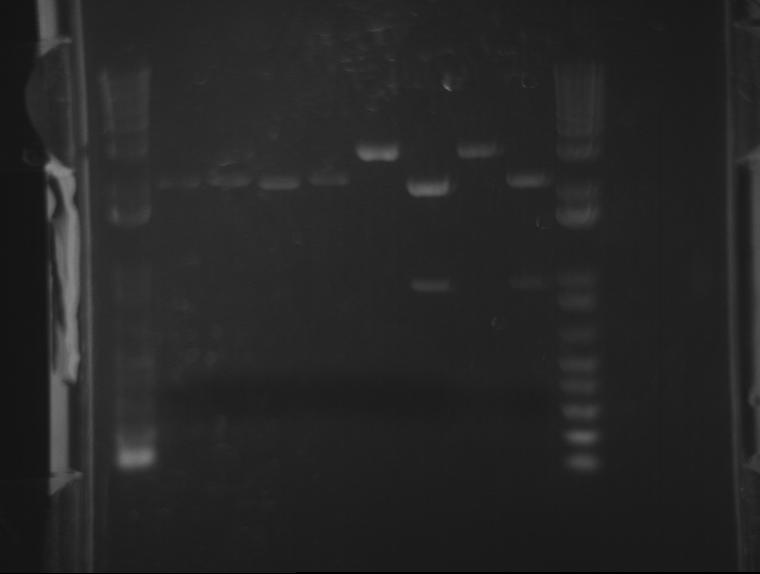
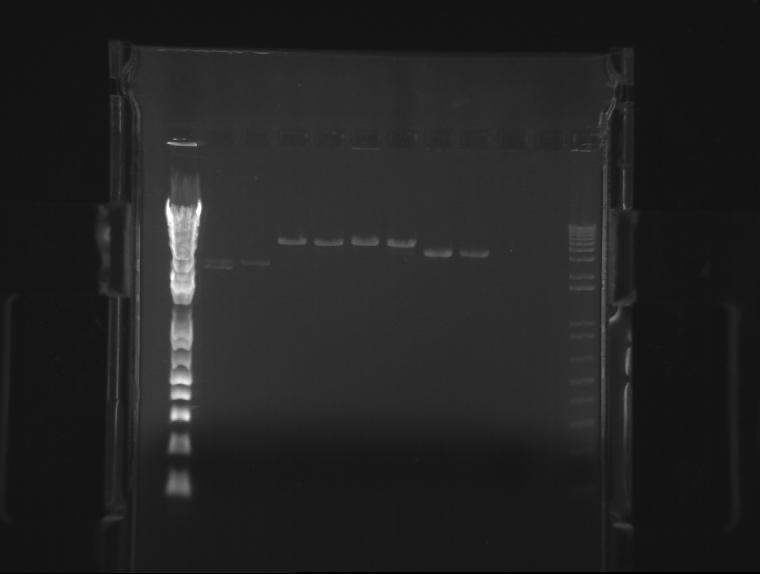
⇒ A 1.4% agarose gel was produced with 0.5% TBE, and 11 samples were run. The gel appears as follows:
The naming scheme proceeds as follows:
L(x)-y indicates ligation number: 4 ligations were produced. See previous posts. L1 corresponds to a 1:1 plasmid/insert ratio, L2 indicates 3:1, etc.
Lx-(y) indicates the square number of the screening plates on which the colony containing the plasmid was found, from square 1 to 25.
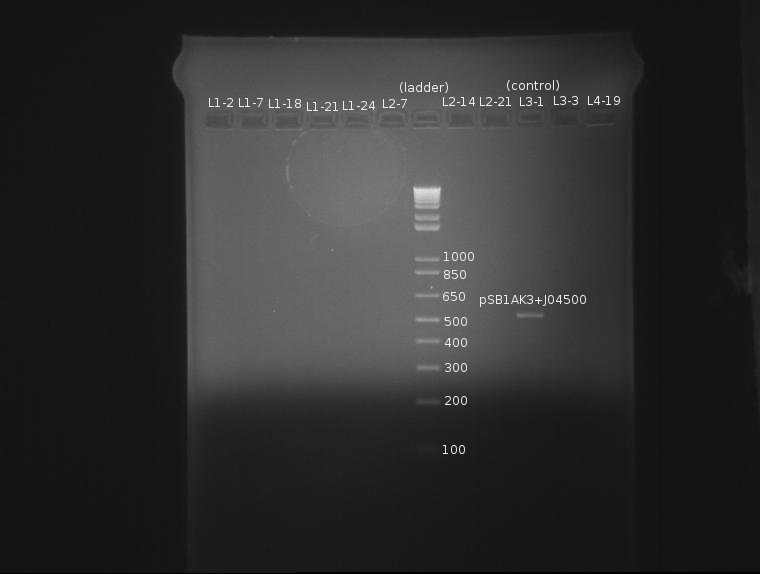
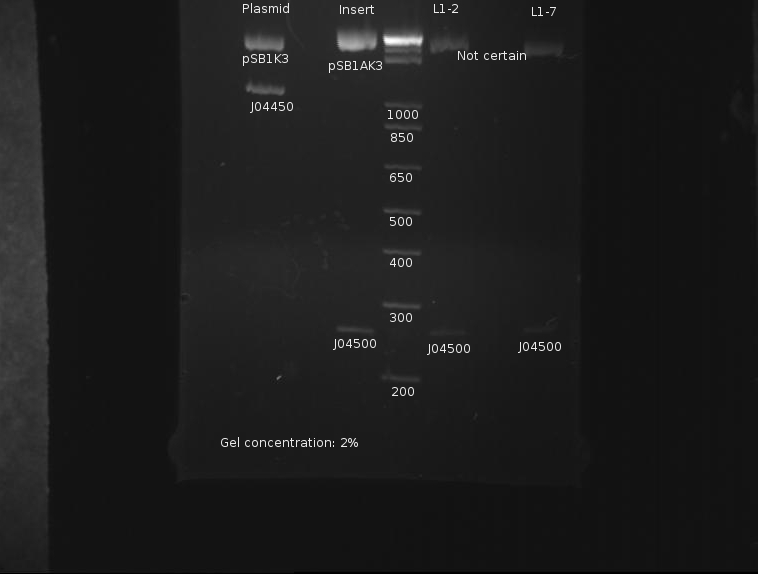
The only bad observed was observed in lane 10, which (I think?) corresponds to the control, pSB1AK3. The lack of any product whatsoever in the other lanes is a disturbing development, as it indicates the failure of the VF2 and VR primers to bind to sites in what is expected to be the pSB1K3 plasmid, or that the plasmid is missing entirely. The fact that all 10 samples are missing bands indicates the problem is likely inherent in the part itself and not due to random lab work error.
⇒ This will be investigated by means of manual overnight cultures. Ligations L1-2 and L1-7 were suspended in 2mL LB+Kan and left to grow. Colonies from plate L1 were chosen for ratio reasons, as the 1:1 plasmid-insert ratio is less likely to have bizzare ligation anomalies.
7/24/2011
Ben
⇒ Overnight cultures were purified using a miniprep kit. A 50µL digestion was prepared containing L1-2, L1-7, and the remains of the two stock parts (pSB1K3 and J04500) as controls. The digestion proceeded as follows:
∴ 32.5µL diH2O
∴ 5µL Buffer
∴ 0.5µL BSA
∴ 10µL DNA solution
∴ 1µL EcoRI and PstI
Incubated at 37°C for 2 hours, then heat inactivated at 80°C for 20 minutes.
⇒ Solutions were first run on a 2% gel to catch and compare the insert, J04500, with the stock parts:
The gel concentration was too high to resolve the plasmids' size. Otherwise, it appears the insert is of correct length.
⇒ Solutions were then run on a separate 1% gel to resolve the plasmid backbones' length:
It is easy to resolve the length of the backbone at around 2200bp here. It is unknown as to why the PCR reaction failed previously, but from this assessment the part appears to now be in the proper backbone.
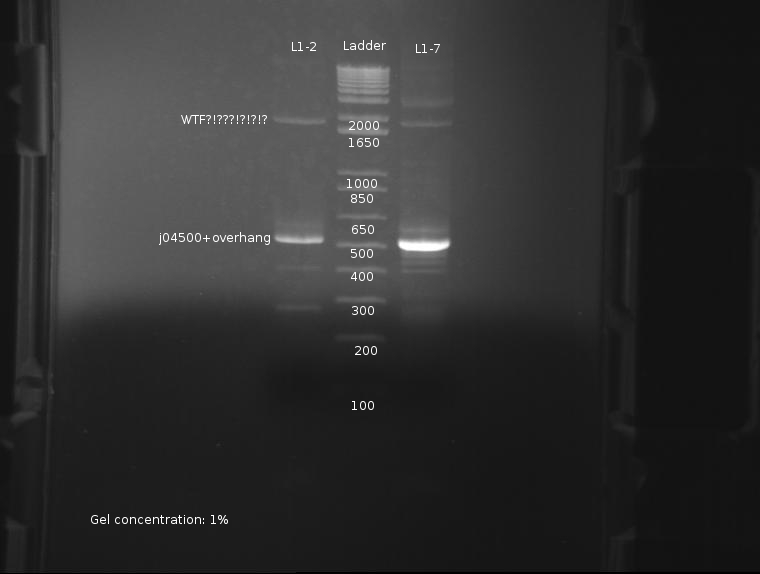
⇒ The PCR reaction was attempted a second time, this time with stocks of purified ligations. The protocol run was identical to the previous reaction. The gel appears as follows:
The strongest band corresponds to J04500 with about 200-300pb of overhang due to the offset locations of the VF2 and VR2 primer sites. This band is of predicted size, further confirming the part's attributes. Worrying, however, are the extra bands seen at around 2000; these bands are present in both ligations, and as such are unlikey to be contamination. It is possible these bands are the VF2/VR primers adhering to terminators and such in the plasmid, as seen here: http://partsregistry.org/Problems_with_PCR_using_VR/VF2
It is currently unknown as to the nature of the failure in the previous PCR reaction, but it likely due to a methodological error, or failure of the usual protocol used, which uses whole cells inserted into the reaction.
7/26/2011
Candy
⇒ Inoculated pSB1K3-J04450, also affectionately nicknamed "pSB1K3", from Ben P.'s plate from earlier (7/19/2011) in order to steal the plasmid to swap Gli-1 into. Gli-1 currently reside in J63009, a low copy plasmid.
7/27/2011
Candy
⇒ Incubate BL21 (generously donated by Ben P.) in 3mL LB
∴ In preparation for -80 degC storage
⇒ miniprep pSB1L3-J04450 from yesterday.
⇒ Double digest with EcoRI and PstI:
pSB1K3-J04450 miniprep (7/27/11)
J63009-Gli1 (6/26/11)
∴In preparation to swap J63009 for pSB1K3 to amplify Gli-1. Haven't figured out if BBa_K165007 (Gli-1) as it was submitted by 2008 Brown(Two) followed RFC-10 or RFC-23. The theory is that the prefix and suffix of Gli-1, whatever it is, will follow the part into pSB1K3.
7/28/2011
Josh
⇒reconfigured digests to include a diversity of parts
Ben
⇒ J257018 (AIDA-I) and J04500(lac+RBS) were inoculated in preparation for a construct with ADIA-I. Additionally, signal peptide part J23015 was removed from the wells and transformed.
7/29/2011
Namun, Ben
⇒ J257018 and J04500 were purified using the Miniprep protocol. J23015 was innoculated in 3mL LB+AMP.
⇒ 100µL of the DH5α overnight culture were innoculated in a second 100mL of LB and left to grow for several hours. When the mixture was barely cloudy, the solution was pelleted. Pellets were very small, so a total of only ~5-10mL of competent cells was produced total. THIS SHOULD BE DONE EVERY WEEK PEOPLE!
⇒ BL21 was innoculated in ~100µL LB in preparation for competent cell production.
⇒ AgeI, NgoMIV, SacI were ordered. NgoMIV is currently on back-order and will arrive later. AgeI and NgoMIV are necessary for the RFC25 protein fusion standard, if we ever want to use it; SacI is useful for cutting the OmpA part.
⇒ To assess the compatibility of Gli-I with the RFC25 standard, a digest was run with EcoRI and AgeI. AgeI is very expensive and should only be used sparingly. The digest showed one distinct, linear band, indicating the lack of an AgeI site. This does not bode well for fusion constructions with Gli-I, although it may be compatible with the RFC23 standard, which uses standard Biobrick enzymes.
7/30/2011
Josh
⇒ Continued reconfiguration of digest.
Ben
⇒ J04500 was digested with EcoRI and SpeI as well as XbaI and SpeI (for Josh.) J32015 was digested with EcoRI and XbaI in preparation to recieve 04500 upstream. However, 04500 was extremely dilute, while 032015 was extremely concentrated. Additionally, SpeI appeared to not have cut completely:
This was a double volume digest of 100µL, which is very bad considering the resources wasted. It may be possible to do 50µL reactions with high concentrations of plasmid and extend the digest, then purify this smaller volume to 30µL in order to save precious enzymes.
Digestion products will probably not be used due to these issues. J032015 was diluted to 50% of its current concentration, as that band is REALLY bright.
7/31/2011
Josh
⇒ Began mixing of digest.
8/1/2011
Ben
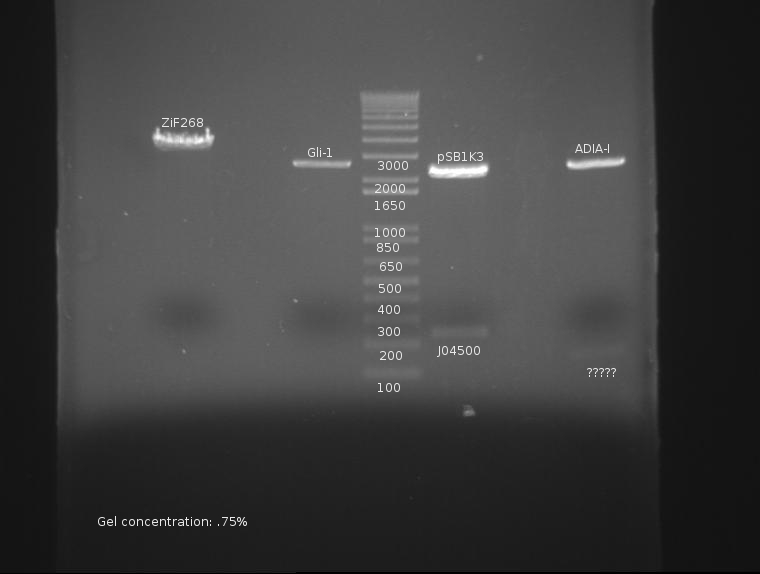
⇒ All components of AIDA-I have been found to be in frame; as such, cloning on this part will continue.
⇒ A giant digestion of ZiF268, Gil-1, ProRBS, and ADIA-I was prepared. The following change(s have been made to this protocol as a test:
∴ 1µL Enzyme instead of 2µL will be used, since a large excess of enzyme is used anyway. This will save enzyme (duh.) This was recommended by Knight in the openwetware protocol.
∴ Digestion time was extended to ~4 hours (up from 2 hours) due to the nature of double digests with SpeI and PstI in NEBuffer 1 (both 75% effective), as well as XbaI and PstI in NEBuffer 3 (XbaI being 75% effective). SpeI was also behaving flakily in the previous digest (see above.)
The digest proceeds as follows:
ZiF268 (K165006): SpeI and PstI in preparation for insertion of ADIA-I (K257018)
Gli-1 (K167007): SpeI and PstI in preparation for insertion of ADIA-I (K257018)
ProRBS (J04500): EcoRI, SpeI in preparation for insertion upstream of J32015
ADIA-I (K257018): XbaI and PstI in preparation for insertion upstream of the zinc fingers.
The gel is as follows:
ADIA-I not only lacks a band at ~1300, but appears to have a faint band at 150. This is very, very bad and may constitute a failure of the part or stock. This will be investigated.
8/3/2011
Chris
1) Miniprepped overnight-ed cultures of ZIF and GLI zinc fingers. Currently stored in -20 freezer.
2) To confirm product, digested the minipreps with EcoR1 and Pst1. Digest solution: 8 microliters miniprep DNA, 1 microliter NEBuffer3, .5 microliters enzyme each.
3) Immediately ran gel of digest upon completion:
8/4/2011
Josh
- completed digest
8/5/2011
Ben
⇒ The past several days have yielded severe trouble with refrigerator stocks of ADIA-I (K257018). The part appears to be bad, as multiple digests with it have yielded a small, 150bp fragment that does not appear anywhere close to the fragment desired in the digest.
⇒ A PCR reaction was planned using small amounts of DNA straight from the wells. This reaction was 20µL in size. The outcome was compared with the digested fridge stock of ADIA-I, which appears to be bad:
The expected part size should be 1320bp, and when the distance between the VF2 and VR sites is taken into account, about 1600. This part is nowhere near these two measurements. For further confirmation, the part will be digested in a 100µL reaction.
This makes sense, but with terrifying implications. Part cut with XbaI (incompletely?) yields two bands; part cut with SpeI also yields two bands; both have cut bands at predicted locations. PCR product cut with both XbaI and SpeI yields a product shorter than both others, indicating the PCR product is not a partial product. The only resonable conclusion(s) at this stage are as follows:
∴ The part is completely wrong, and was put in the wrong well (and is hence unusable.)
∴ The VF2 and VR primers somehow generated product by binding to completely wrong locations on the plasmid, while still producing predictable products.
When viewed in an earlier snapshot, bands around 150-170bp are notably missing; these would correspond with subsequent "tail pieces" cut off of the part by the restriction enzymes. Unfortunately, the sample appears too dilute and the cutting too incomplete if conclusions can be drawn from this missing component.
⇒ The last of AIDA-I (K257018) was transformed and plated from residue in the wells in the hopes overnight cultures + digestions will be able to resolve this matter conclusively.
⇒ A ligation of J32015 and J04500 was completed using previously digested stocks. Three ligations total were produced using the standard ligation protocol. Ligation products were plated on AMP plates.
8/6/2011
Ben
⇒ Colony PCR was run on 6 ligation samples, as the ligation of SP (J32015) and the promoter/rbs (J04500) appears to have grown properly on the plates. Also in the PCR were colonies from the elusive ADIA-I part as well as J04450 from the RFP colonies as a control.
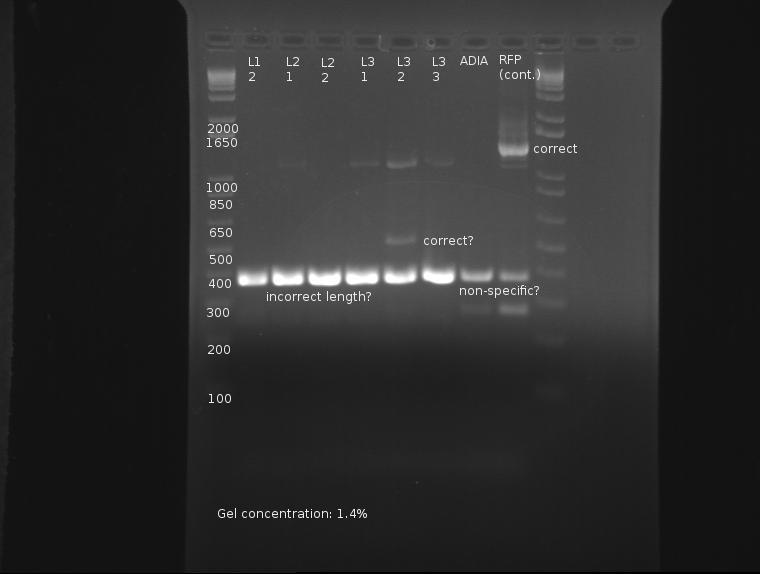
This gel is difficult to interpret. All ligation bands appear to be around 360-380bp in length, which, unfortunately, corresponds with J32015 without J04500. While an indication the ligation failed, the presence of these bands in both the AIDA-I part (which failed to have any other bands) as well as the RFP part are strange and may indicate nonspecific binding. Since no control was run on the SP stock itself as verification, this gel is not conclusive enough to conclude on the state of the ligation.
⇒ A second colony PCR was set up containing six samples; this will finish tomorrow. Ligations L3-1 through L3-3 will be compared with PCR run on stocks of J04500, J32015, and J04450 as controls.
⇒ Ligations L3-1 through L3-3 were also innoculated in 3mL LB+AMP, as well as ADIA as an overnight. All these will be purified and digested to determine the true nature of their inserts.
8/7/2011
Ben
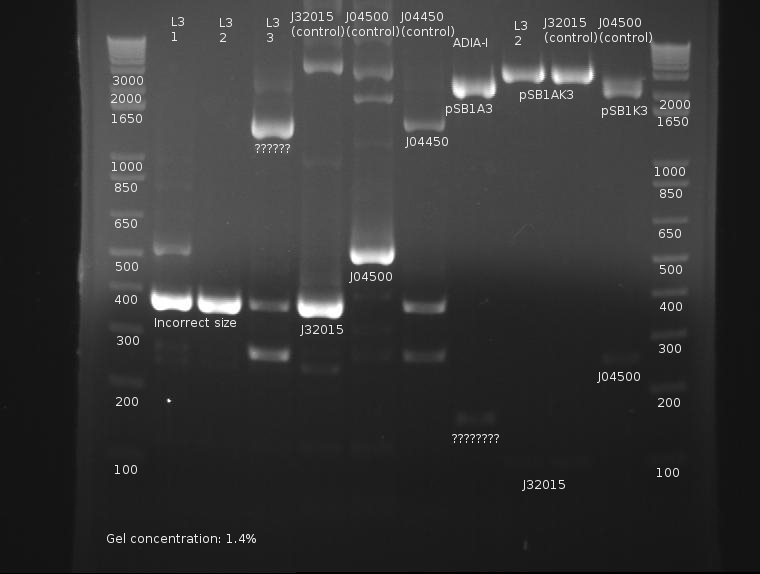
⇒ Overnights of L3-1,L3-2,and L3-3 were checked. Strangely, only L3-2 possessed any growth at all. ADIA-I also possessed growth. These were miniprepped and digested with EcoRI and PstI in a 50µL reaction mixture. These were run on a gel along with the 6 sample PCR from yesterday.
Extreme nonspecific binding aside, the ligation is confirmed as failed. Bands in both the PCR and digestion of cultured products indicate the screened ligations are meerly the signal peptide (J32015) in its native backbone, with no ligated material.
⇒ The VF/VR PCR program has been modified to be in-line with the recommended NEB protocol for Phusion polymerase.
⇒ Comp cells were produced and now reside in the refrigerator.
8/09/2011
Alena
⇒ Miniprep 2 mL of both His-tagged ZFPs (His-Gli1-nCFP: BBa_K323058 and His-Zif268-cCFP: BBa_K323069) from 8/04/11
∴ for BL21 transformation
Candy
⇒ The University of Michigan Sequencing Core delivered sequencing results for BBa_K165006 (Zif268-HIV) and BBa_K165007 (Gli-1) using either VF or VR primer, diluted to 1 uM, that we provided along with the miniprep samples:
∴ Gli-1 with VF primer had RFC23 prefix and suffix, and the part itself is perfect match with partsregistry information.
∴ Results of Zif268-HIV with VF may be showing a part of the backbone or something, as nothing remotely like the part appeared, but the reverse compliment of sequencing with VR had a near perfect RFC 23 prefix (T->A immediately after NotI site) and perfect RFC 23 suffix, the part itself is a 98% match with what's in partsregistry and in the right place. Haven't checked implication in translation yet.
∴ Neither ZFP had sequencing information in "get this part".
8/10/2011
Alison, LJ, Kevin
⇒ The 3 of us from the Printing group underwent our protocol to test the blank slides sent to us from Mycroarray. (see protocols for more details). We tested 3 different blank slides: one with DH5alpha cells, one blank, and one with cells and a dried milk "blocking" solution meant to keep the cells from sticking to the slide. After completing our protocol (adding a rinse with Tween 20 and multiple rinses of PBS), we studied the 3 different slides under a microscope and compared their similarities and differences.
⇒ The blank slide featured some spots that we think were dirt, as there were no cells in contact with this slide. The cell slide obviously had E Coli cells stuck to it of different shapes and sizes. The shapes were circular, like the dirt on the blank slide, but were definitely larger. The slide with the "blocking" solution was interesting in that it had very little attached to it; it succeeded in preventing cells from sticking to the glass. However, we did notice what must have been a piece of dried milk stuck to the slide. It looked different than anything else we saw underneath the microscope and seemed similar to a piece of muscle or the blood vessels in the lungs. We will remember that for the future if we use the blocking solution.
Ben
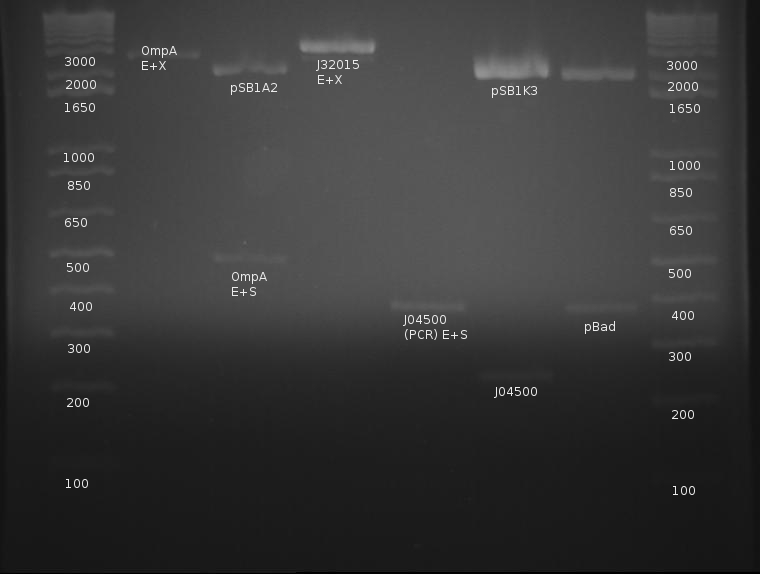
⇒ PCR was run on pBad (K259007) and lac (J04500) in preparation for their respective insertions upstream from OmpA. A 5prime master mix was used, along with 1 microliter of VF2 and VR primers.
8/11/2011
Ben
⇒ Ompa (K103006) was digested with EcoRI and XbaI in a 100μL reaction; the reaction was held for ~3 hours, then inactivated at 80°C for 20 minutes.
⇒ All three components were purified and run on a gel along with a signal peptide (K32015) digestion from like a week ago:
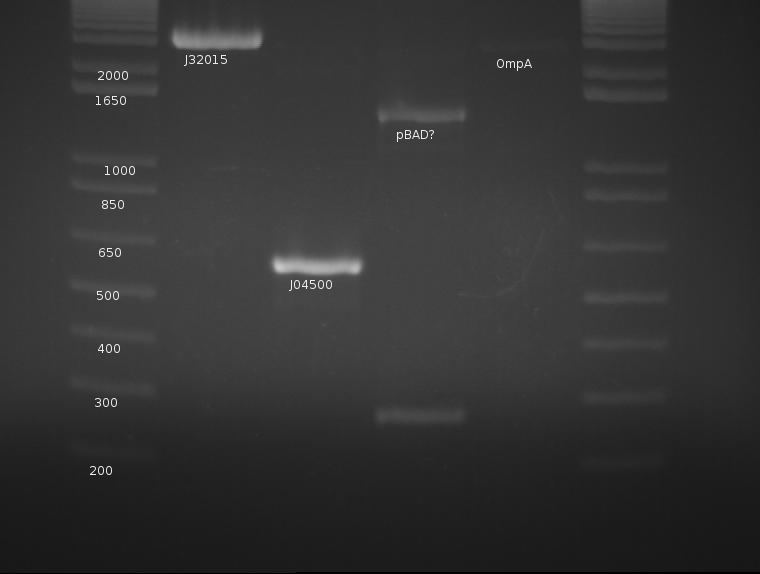
OmpA appears extremely faint, likely due to the failure of the stock solution used.. J04500's PCR worked very well and yields a strong band with little to no mispriming; this will be cut later with EcoRI and SpeI. pBad is a mystery, as neither band corresponds to the correct length of the part. The signal peptide (J32015) is highly concentrated and appears well cut; this can proceed with the ligation tomorrow. pBad is generating two creepy PCR bands and will be discarded.
⇒ To continue, J04500 was cut with EcoRI and SpeI in a 3 hour, 50μL as it was quite concentrated. A gel was run with the result of the digestion compared with a previous, cut stock (less concentrated) as well as the raw PCR product.
⇒ Overnights of the signal peptide, pBad and OmpA were prepared for the hopeful ligation tomorrow.
Josh
Gel 1:
1 - ladder
2 - RBS J04500 EcoR1 + Spe1 (3189 bp + 220 bp)
3 - RBS J04500 Spe1 + Pst1 (3409 bp)
4 - INP + Backbone EcoR1 + Xba1
5 - INP + Backbone EcoR1 + Spe1 (960 bp)
6 - INP + Backbone Xba1 + Pst1 (960 bp)
7 - INP + Backbone Spe1 + Pst1
8 - INP 2 Xba1 + Pst1 (960 bp)
9 - INP + linker PCR EcoR1 + Spe1 (960 bp)
10 - OmpA (K103006) Xba1 + Pst1 (464 bp)
Gel 2:
1 - ladder
2 - AID-A EcoR1 + Xba1
3 - AID-A Xba1 + Pst1
4 - AID-A 2 EcoR1 + Xba1
5 - AID-A 2 Xba1 + Pst1
6 - linker 1 EcoR1 + Xba1
7 - linker 2 Spe1 + Pst1
8 - constitutive promoter/RFP 1 EcoR1 + Spe1
9 - linker + GFP 1 EcoR1 + Xba1
10 - linker + GFP 1 Xba1 + Pst1
11 - linker + GFP 2 Xba1 + Pst1
12 - added duplicate lane somewhere (redo the gel)
8/12/2011
Ben
⇒ OmpA, signal peptide, lac and pBad stocks were purified from their overnight form. All stocks yielded poor growth, with OmpA being the lowest yield.
⇒ Competent cells were produced from a BL21 DE3 overnight of unknown origin using a protocol from the Jakob lab. This protocol is not currently available.
⇒ OmpA was digested with EcoRI and XbaI, as well as EcoRI and SpeI for verification. The signal peptide was digested with EcoRI and XbaI to prepare for an upstream insertion. pBad, being an insert, was digested with EcoRI and SpeI. A gel was run to confirm as follows:
OmpA was digested with E and S as a confirmation of its length. From this, a band can be seen around 480-500, which is correct length. The signal peptide cannot be confirmed about the completion of its digestion; this is also the case with OmpA cut with E and X. pBad appears to have cut correctly, and the previous PCR failure is unknown in cause. What is disturbing, however, is the cut length of the lac promoter, J04500.
Repeated experiments with this indicate either only one side of the PCR product is being cut, or the part is incorrect in its entirety. Previous gels indicate J04500 may be in fact too long to be the correct part, as PCR run on this part shown in previous gels appears slightly too long to begin with.
After repeated failures of PCR product in this part, something may be wrong with our protocols. This will be tested over the next week or so.
8/14/2011
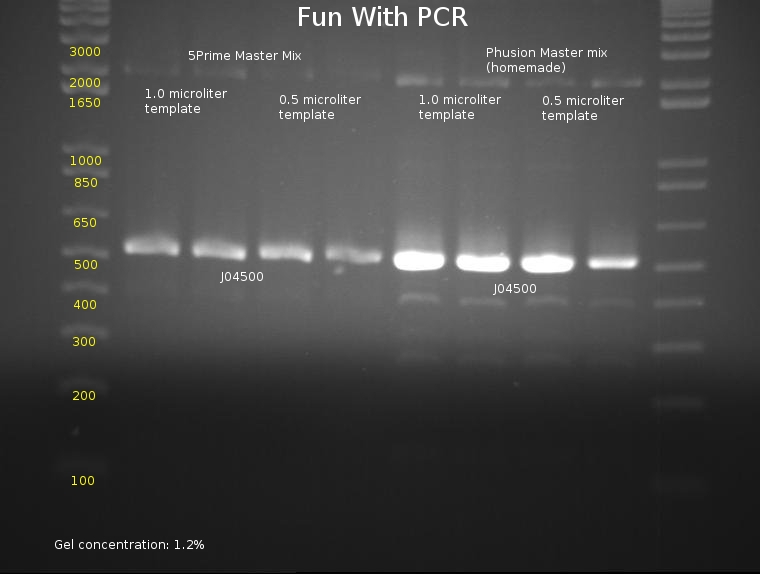
Ben
⇒ An 8-lane PCR is being tested on J04500 using Phusion mix and 5prime Master Mix. The PCR will test for reproduciblity and ultimately hope to determine the best methods of PCR, as the last series of (fragmented) PCRs have proved to be extremely inconsistent. The following mixtures were used:
5Prime MM
∴ 40µL 5Prime master mix
∴ 35µL diH2O
∴ 10µL VF2 (diluted to 1µM)
∴ 10µL VR (diluted to 1µM)
Phusion MM (home-grown from NEB website protocol)
∴ 20µL HF Buffer (5x)
∴ 2µL dNTP solution
∴ 5µL VF2 (stock)
∴ 5µL VR (stock)
∴ 1µL Phusion polymerase
⇒ An gel was run with these components to assay PCR reliability:
Frustratingly, all bands appear to be at normal locations. Reasoning for previous PCR failures are unknown; possible errors include bad stock (which was changed for this assay) or pipetting errors generating unpredictable product. Admittedly, A different template stock was used for this assay then previous weekend experiments, which may be a component of the problem.
Phusion polymerase obviously is far more powerful than the 5Prime master mix; at the same time, it must be noted it costs almost 5 times as much. Little remains of the stock in the freezer, so, due to cost restraints, using the 5Prime master mix is our best bet unless you actually plan to mass-produce a part for the purposes of cloning it. Before producing these parts, assaying their behavior with PCR using 5Prime MM first is a good idea. 5Prime MM should be used for all colony PCRs and all options where extremely high concentrations of highly precise product are not needed.
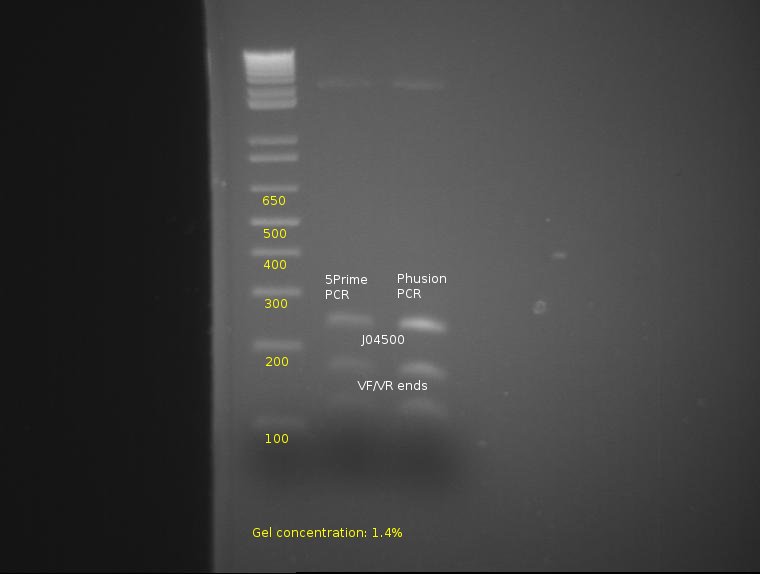
⇒ To continue the assay, the four lanes corresponding to 5Prime and Phusion were combined, purified, and are being cut with E and S to determine prefix/suffix integrity.
From this, proper bands appear at 260, 170, and 120, indicating the parts have been properly cut and possess correct integrity from the PCR.
8/15/2011
Ben
⇒ J04500 was ligated into OmpA and SP plasmids; both were plated on AMP. Colony PCR will be attempted in the morning.
8/16/2011
Ben
⇒ Colonies from the ligation were observed, but were few and far between. They will be grown for ~18 hours before colony PCR is attempted.
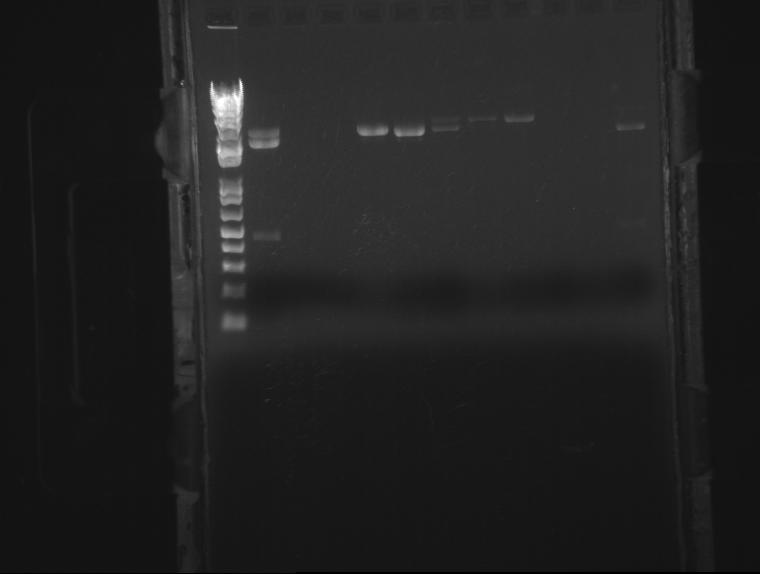
⇒ PCR was run on a Gli-OmpA hybrid part. Unfortunately, stocks were confused and this part turned out to be OmpA. PCR was re-run on a confirmed correct part, for use in assembly with J04500 via 3a construction.
If all goes according to plan, PCR should become an integral component of cloning efforts. Inserts can be produced in high concentration with no backbone to screen against in 2-3 hours, instead of 16 hour overnights, and purified in 20 minutes instead of miniprepping. Higher concentration of backbone can be used for the higher concentration required for 3a ligation.
Josh and Amy
⇒ configured new ompA, OmpA-gli1, gli1, ZiF, and ZnF268 digests
8/17/2011
Ben
⇒ PCRs of OmpA-Gli and J04500, along with a miniprep of pSB1K3+J04450, were digested in preparation for 3a construction and purified. The gel, while a mess, still indicates proper cutting of the VF/VR extension artifacts.
 "
"