Team:KAIST-Korea/Projects/report 42
From 2011.igem.org
(Difference between revisions)
| Line 52: | Line 52: | ||
[[File:Fig1-report4.png|800px|thumb|center| Fig 1. (a)schematic representation of light detection. In order to detect light, minimum visual acuity and light intensity should be satisfied. (b) Quantitative information of photon emission by a GFP molecules]] | [[File:Fig1-report4.png|800px|thumb|center| Fig 1. (a)schematic representation of light detection. In order to detect light, minimum visual acuity and light intensity should be satisfied. (b) Quantitative information of photon emission by a GFP molecules]] | ||
| - | :We assume that the E. coli is in a darkroom for discovering the minimum number of fluorescent protein. In this reason, we use 0.1 lx, the minimum intensity of light that cone cells in human can perceive. [2] In fig 1 (a), There is a brief picture for minimum visible acuity. We choose that 1 second is the basic time scale, so have to know the number of fluorescent protein per 1 second. By our research, The range of the photon emitted time is wide, from <math>10^-5</math>s to <math>10^-8</math>s Although it hasn’t any theoretical reason, we choose the photon emitted time by a fluorescent protein, .[3] | + | :We assume that the E. coli is in a darkroom for discovering the minimum number of fluorescent protein. In this reason, we use 0.1 lx, the minimum intensity of light that cone cells in human can perceive. [2] In fig 1 (a), There is a brief picture for minimum visible acuity. We choose that 1 second is the basic time scale, so have to know the number of fluorescent protein per 1 second. By our research, The range of the photon emitted time is wide, from <math>\10^-5</math>s to <math>10^-8</math>s Although it hasn’t any theoretical reason, we choose the photon emitted time by a fluorescent protein, .[3] |
:We will proceed this modeling by using the energy of photons for relating the light and the number of fluorescent protein. One of the unit for light is lm(lumen) and this unit is transformed to J(joule) in 1 lumen. | :We will proceed this modeling by using the energy of photons for relating the light and the number of fluorescent protein. One of the unit for light is lm(lumen) and this unit is transformed to J(joule) in 1 lumen. | ||
Revision as of 07:14, 7 August 2011
Fluorescence Visibility Justification
The human eyes cannot perceive objects that are smaller than a certain size. Also, they cannot recognize light whose intensity is lower than an inherent threshold. We take these limitations into account to determine the number of fluorescent proteins that must accumulate before we can notice any fluorescence, and establish the minimum circular area required for us to perceive any fluorescence. Therefore, the objective of this modeling is following:
To investigate concentration required for an amount of fluorescent protein in the E.coli that makes light from E.coli be visible for human.
Background
- Human has the limits in vision. For our objective, we have to know about the limit of recognizing size of objects in human vision. This limit is called the ‘Minimum visible acuity’. The exact definition of minimum visible acuity is the minimum size of object that the human eyes can discern. In Table 1. Types of visual acuity[1], the value of detection acuity(red box), ~1.0 arc second, is the minimum visible acuity that we take.
- We assume that the E. coli is in a darkroom for discovering the minimum number of fluorescent protein. In this reason, we use 0.1 lx, the minimum intensity of light that cone cells in human can perceive. [2] In fig 1 (a), There is a brief picture for minimum visible acuity. We choose that 1 second is the basic time scale, so have to know the number of fluorescent protein per 1 second. By our research, The range of the photon emitted time is wide, from <math>\10^-5</math>s to <math>10^-8</math>s Although it hasn’t any theoretical reason, we choose the photon emitted time by a fluorescent protein, .[3]
- We will proceed this modeling by using the energy of photons for relating the light and the number of fluorescent protein. One of the unit for light is lm(lumen) and this unit is transformed to J(joule) in 1 lumen.
Program input
- The user provides the program with the concentrations of the two types of E.coli, one that produces quorum and another that responds by producing fluorescent proteins, the dimensions of the medium, which in this case serves as the canvas, and a sketch of the picture by either directly moving the cursor around on the screen (intentional seeding) or by employing the random seeding function which randomly scatters both types of E.coli across the canvas (random seeding).
- Each method of seeding displays distinct characteristics. As mentioned earlier, random seeding randomly scatters both types of E.coli across the canvas, which results in most cases a conglomeration of colors uniformly spanning the canvas. On the other hand, intentional seeding allows the user to draw specific figures or delineate curvatures as he wishes. Because the amount of E.coli dropped onto the canvas is proportional to the time until the mouse is unclicked, he can choose to seed heavily or lightly by controlling the mouse appropriately. Lastly, the higher the concentration provided to the program, the heavier the default seeding.
Algorithm
- In each frame, the following series of steps take place. First, Brush E.colis produce quorums, which diffuse to adjacent cells. As established earlier, quorum diffuses sufficiently fast to adjacent cells that we can guarantee it diffuses to neighboring cells well before the advent of the next frame. Upon receiving quorum, paint E.colis produce quorum and fluorescent proteins, whose expression is modeled in our program as solid RGB colors occupying the area of each cell. The previous series of steps constitute all the components contained within a frame. The process repeats every minute, which is a repetition of the same process described here.
Output
- The program will save and display the resultant randomly generated color picture over a user-specified time period. It will also store the progress of the effect of quorum sensing and fluorescent protein production in discrete, chronologically consecutive frames. It also saves and displays the distribution of each type of E.coli across the canvas.
Results & Analysis
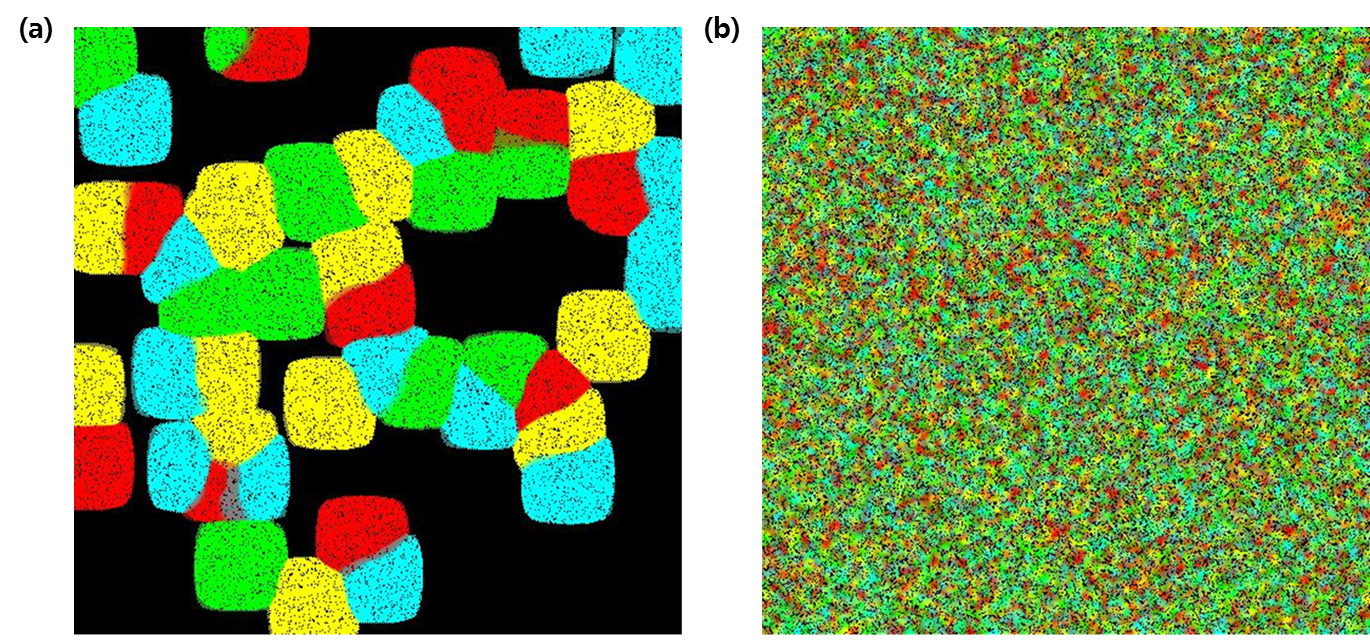
- Figure 2 displays the paintings obtained by randomly seeding 50 Brush E.coli and 200,000 paint E.coli (Fig 2. a) and 30,000 Brush E.coli and 150,000 paint E.coli (Fig 2. b) across the canvas. We obviously observe a difference in the density and uniformity of colors between the two pictures. In the Fig 2. a, distinct colors are clearly visible as Brush E.colis outnumbered the paint E.colis; borderlines between colonies expressing the same colors are clearly delineated. On the contrary, the painting on the right is chaotic; there are no clear colors visible but instead the entire canvas is a uniform mixture of the four colors. Of course, this is expected since the number of E.coli opting to “expand its own colorful empire” is now comparable to that of the paint E.coli.
- Figure 3 shows the result of intentional seeding. In the left, the user moved and clicked the mouse in the shape of iGEM. In the right, KAIST was written. The distributions of each type of E. coli are displayed above each painting.
- If you want to see more detailed results, check out the <'Simulation Results Gallery'>
 "
"