Team:Paris Liliane Bettencourt/Notebook/2011/08/03/
From 2011.igem.org
(→LacI:GFP 37°C) |
(→LacI:GFP - Lac O array) |
||
| Line 48: | Line 48: | ||
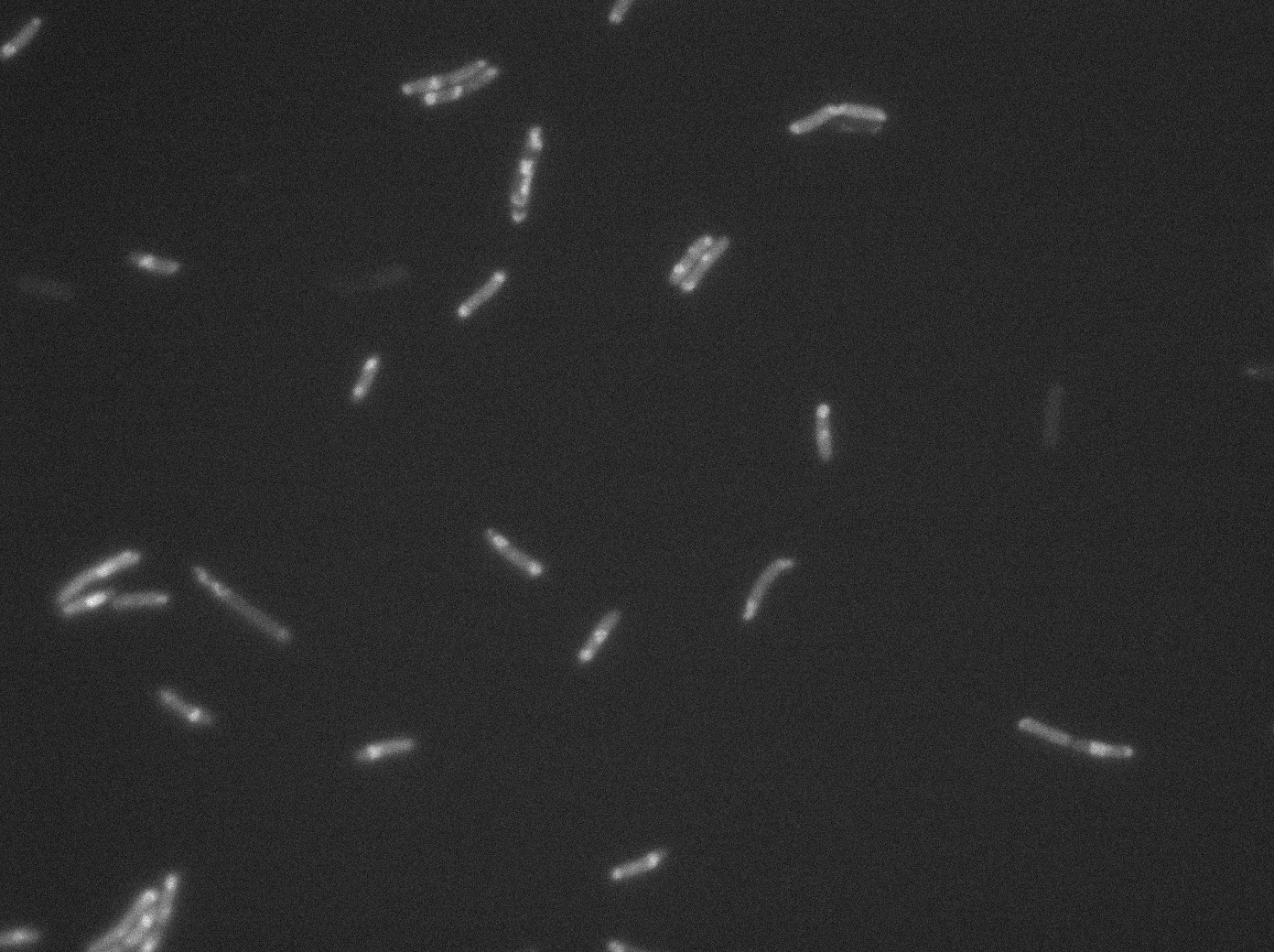
====LacI:GFP - Lac O array==== | ====LacI:GFP - Lac O array==== | ||
| - | [[File:Lac0_Trans03.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with no arabinose on E. Coli from Dave Lane plasmids.]] | + | |
| - | [[File:Lac0_GFP03.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with no arabinose on E. Coli from Dave Lane plasmids.]] | + | {| border="1" class="wikitable" style="text-align: center;" |
| - | [[File:Lac0.1_Trans02.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,1% arabinose on E. Coli from Dave Lane plasmids.]] | + | |+LacI:GFP /LacO array : 37°C |
| - | [[File:Lac0.1_GFP02.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,1% arabinose on E. Coli from Dave Lane plasmids.]] | + | |- |
| - | [[File:Lac0.2_Trans01.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,2% arabinose on E. Coli from Dave Lane plasmids.]] | + | |[[File:Lac0_Trans03.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with no arabinose on E. Coli from Dave Lane plasmids.]] |
| - | [[File:Lac0.2_GFP01.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,2% arabinose on E. Coli from Dave Lane plasmids.]] | + | |[[File:Lac0_GFP03.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with no arabinose on E. Coli from Dave Lane plasmids.]] |
| + | |- | ||
| + | |[[File:Lac0.1_Trans02.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,1% arabinose on E. Coli from Dave Lane plasmids.]] | ||
| + | |[[File:Lac0.1_GFP02.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,1% arabinose on E. Coli from Dave Lane plasmids.]] | ||
| + | |- | ||
| + | |[[File:Lac0.2_Trans01.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,2% arabinose on E. Coli from Dave Lane plasmids.]] | ||
| + | |[[File:Lac0.2_GFP01.jpg|350px|thumb|center|GFP:LacI / LacO array inducted with 0,2% arabinose on E. Coli from Dave Lane plasmids.]] | ||
| + | |} | ||
==Danyel & Camille== | ==Danyel & Camille== | ||
Revision as of 10:14, 4 August 2011
Contents |
Cyrille
Going on with the QCM experiments
The transformation have worked pretty well. The negative control before digestion has some clone, and the negative control after digestion has none. On all the plate, we have a important number of colony, more important than in any negatives control. This indicate that the PCR may have worked.
On the selected clones, the last two (11, and 12) comes from very small colonies on the plate. The hypothesis is that they might be parasite, or clones grown with a non metilated plasmid. Anyway, the yield obtained after the miniprep is poor, and they may come from parasite.
The miniprep was done over 12 selected clones. The culture was grown overday, started at 9:30 and centrifugated at 15:00. The yields are around 500ng/µL depending on the the tubes. The concentration was measured with the Tecan machine.
1µg of each of the 12 clones (except for the last two, from which we took the maximum quantity that is to say 8 µL) was digested using EcoRI-Fast digest in green digestion buffer, for 15 min at 37°C. The result was loaded on the gel, and let run for 20 min at 50V and then at 100V for the rest. Here is the result of yhe gel:
The result is again negative. There are two bands at 3kb indicating the plasmid still have two restriction sites.
We will start again tomorow with Ariel menages digestion protocol...
Kevin
Culture of double transformed cells (suite)
Diluted at 1/100 => expected to be between OD 300 and 600
- 3 Tubes pSG20 at 37°C
- 3 Tubes pFX234 at 37°C
- 3 Tubes pSG20 only at 30°C
- 3 Tubes FX234 only at 30°C
- 3 Tubes pSG20/pDAG464 at 30°C
- 3 Tubes pFX234/pDAG479 at 30°C
At OD 300-600, induction of fusion protein with arabinose 0%, 0,1 % and 0,2% for 1h30.
 "
"