Team:Wageningen UR/Project/ModelingProj1
From 2011.igem.org
(Difference between revisions)
(→Mathematical approach) |
(→Mathematical approach) |
||
| Line 15: | Line 15: | ||
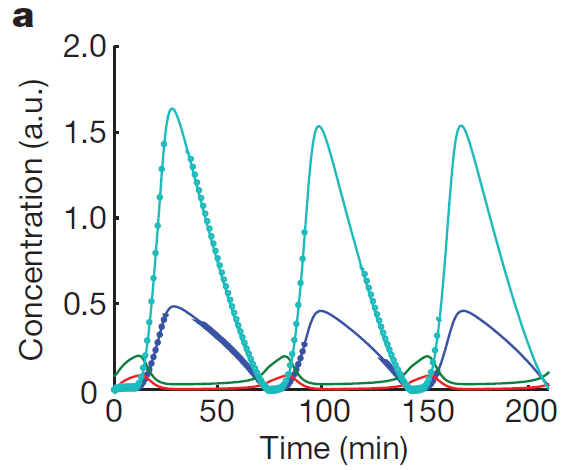
| - | [[File:Template.png|center| | + | [[File:Template.png|center|300px|Graph from the paper "A synchronized quorum of genetic clocks", Tal Danino, Octavio Mondragón-Palomino, Lev Tsimring2 & Jeff Hasty, Nature, January 2010]] |
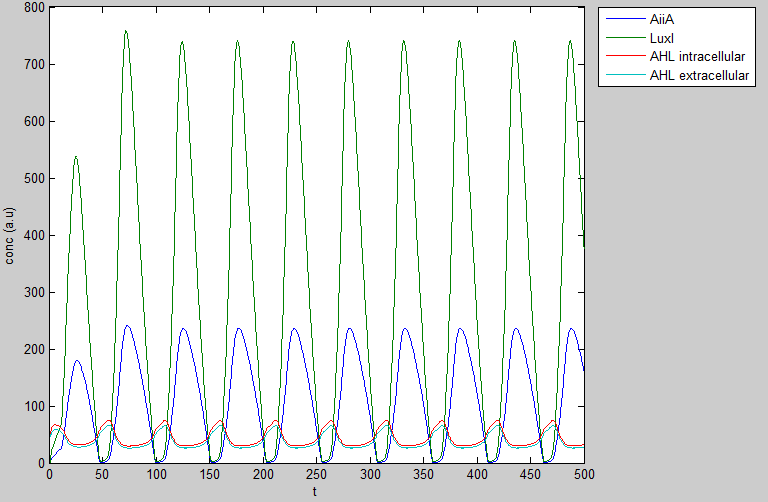
The graph was created using 4 delay differential equations, which were plotted in matlab by the modeling team, obtaining figure 2 as shown below. | The graph was created using 4 delay differential equations, which were plotted in matlab by the modeling team, obtaining figure 2 as shown below. | ||
| - | [[File:Own_WUR.png|center| | + | [[File:Own_WUR.png|center|300px|Reproduction of the oscillatory model done in MATLAB.]] |
Revision as of 17:20, 10 May 2011
Modeling the synchronized oscillatory system
Biological approach
Mathematical approach
Our first approach towards the modeling of our synchronized oscillatory system was to reproduce the model used by Danino et al. in the article "A synchronized quorum of genetic clocks", published in Nature in January 2010. This model will be further adapted to fit the specific devices and ressources available for the Wageningen UR iGem Team 2011. Figure 1 shows theoretical oscillation obtained by the authors of the paper and depicts "A typical time series of concentrations of LuxI (cyan circles), AiiA (blue circles), internal AHL (green line) and external AHL (red line)."
The graph was created using 4 delay differential equations, which were plotted in matlab by the modeling team, obtaining figure 2 as shown below.
 "
"