Team:Peking S/project/wire/inverter
From 2011.igem.org
Nuanuan128 (Talk | contribs) |
Nuanuan128 (Talk | contribs) |
||
| Line 71: | Line 71: | ||
We selected rhl-box from the promoter region of rhl-specific gene rhlA, to which LasR (another transcriptional regulator in Pseudomonas aeruginosa) doesn’t bind, and then positioned the rhl-box inside an artificial lacZ promoter as illustrated in Fig 4, between and partially overlapping the consensus -35 and -10 hexamers, RNA polymerase binding sites. We also replicated work done by Kristia Egland et al which serves as a positive control of our design. | We selected rhl-box from the promoter region of rhl-specific gene rhlA, to which LasR (another transcriptional regulator in Pseudomonas aeruginosa) doesn’t bind, and then positioned the rhl-box inside an artificial lacZ promoter as illustrated in Fig 4, between and partially overlapping the consensus -35 and -10 hexamers, RNA polymerase binding sites. We also replicated work done by Kristia Egland et al which serves as a positive control of our design. | ||
| - | <center> | + | <center>[[File:nuanuan3.png|450px]]</center> |
| - | [[File:nuanuan3.png|450px]]</center> | + | |
''Figure 4: (A) Building Rhl repressible promoter interfaced with appropriate chemical wires. (B) The overall architecture of our quorum sensing repression system.'' | ''Figure 4: (A) Building Rhl repressible promoter interfaced with appropriate chemical wires. (B) The overall architecture of our quorum sensing repression system.'' | ||
Latest revision as of 14:27, 22 November 2011
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Chemical Wire Toolbox
Introduction|Harvesting ‘Chemical Wires’ From Nature|Synthesizing Quorum Sensing Inverters|Orthogonal Activating Matrix
Synthesizing Quorum Sensing Inverters
Inverter is one of the basic genetic parts that need to be developed. It can be anticipated that the absence of feedback mechanisms in Boolean or Non-Boolean gene networks is definitely a major hurdle towards complex behaivor for synthetic microbial consortia.
All of the quorum sensing systems currently exploited in synthetic biology exhibit transcriptional activation, which cannot provide negative feed back loops during cell-cell communication. Therefore, a switch that produces a negative feed back, an inverter in another word, was conventionally implemented by linking an input promoter to the expression of a repressor, which then turns off a downstream promoter generally, using the repressor-operator pairs as shown in Fig 1.
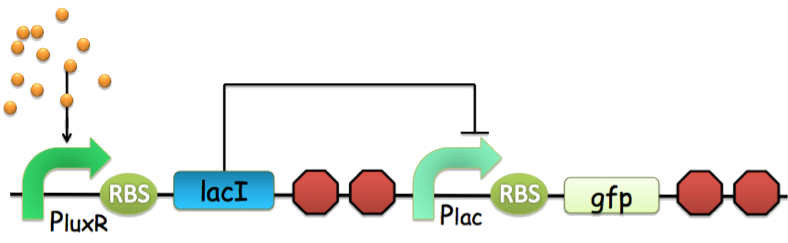
Figure 1: An archetype of conventional genetic inverter. The core component of this inverter is a repressor-operator pair (in this case, the lacI-LacO pair), which requires a transcription-translation delay process of nearly 40 minutes.
However, this two-node repression architecture has several defects. First, the indirect repression would lead to the propagation of metabolic burden and noise through networks comparing with direct repression. Furthermore, multi-component transcriptional cascades can result in a significant time delay. It is generally recognized that a one-stage cascade reaches its half-maximal activation in minutes, whereas for a three-stage cascade this may cost several hours. In some applications, for instance, biosensors for environment detection or disease diagnosis, even a ten-minute delay may be too long. What’s more, it is often difficult to synthesize inverters under certain principles since two parts cannot be connected in series when their timing and dynamic range do not match. In this sense, re-building inverter by fundamental components is necessary.
Kristia Egland and E. P. Greenberg revealed that a quorum-sensing negatively regulated promoter could be constructed. There a luxR dimer could behave as a repressor when binding to its cognate effector molecule, AHL (acetyl-Homoserine Lactone). They created this artificial promoter by positioning the lux-box between and partially overlapping consensus -35 and -10 hexamers with 18 bp between the hexamers. When AHL represents and binds to luxR proteins, luxR will bind to its DNA binding site, the lux box, thus to reduce the accessibility of RNA polymerases to this promoter, repressing the transcription initiation.
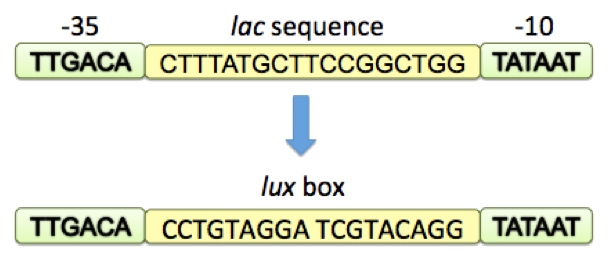
Figure 2: Building lux repressible promoter with lux box. This promoter was built by positioning the 18-bp lux box between and partially overlapping consensus -35 and -10 hexamers.
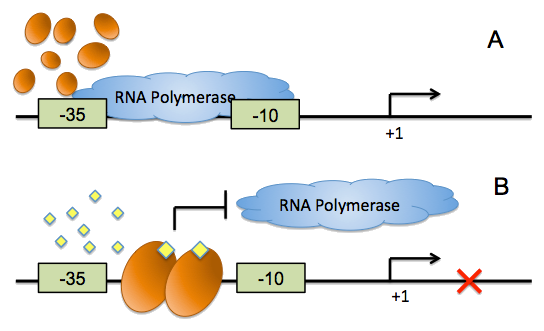
Figure 3: The mechanism of AHL repressible promoter. (A) RNA polymerase functions well when AHL signal is absent. (B) When AHL presents and binds to transcriptional regulator, the transcription regulator binds its DNA binding site, thus to reduce the accessibility of RNA polymerase to promoter, repressing the transcription initiation.
This work inspired us that harnessing the conditioned binding of quorum sensing regulators to their cognate DNA box to control the accessibility of RNA polymerase would be a way of general interest to design direct, fast and reliable signaling inverters for synthetic microbial consortia.
In fact, a significant aspect of synthetic biology is the re-design of existing, natural biological systems for useful purposes. Applying rational design to parts alteration is advantageous, in that it can not only generate products with defined function, but also produce biological insights into how the designed function comes about.
We applied this framework to develop a method to synthesize quorum-sensing inverters. This method permits us to systematically construct promoters in a modular fashion, as well as provides us with an experimental foundation that can be utilized to develop thermodynamic models in which input (the concentrations of chemical signal) and output (the gene repression fold) can be computationally predicted and explained. Inverters under the same architecture reasonably share most common features in regulation model.
Designing quorum-sensing repressible promoter from ground up
We selected an AHL-based quorum-sensing system, RhlR-RhlI pair from Pseudomonas aeruginosa as a proof of concept to design quorum sensing repressible promoter. RhlI is a synthase that produces signal molecule butanoyl (C4-HSL) and RhlR is the regulator that responds to C4-HSL, supposed to activate transcription at cognate rhl promoter. Genetic evidence suggests that RhlR binds to conserved sequence of some quorum-controlled promoters. This so-called rhl box-like sequence shows conserved homology to the lux-box, which is a promoter element required for quorum control of the Vibrio. fischeri.
We selected rhl-box from the promoter region of rhl-specific gene rhlA, to which LasR (another transcriptional regulator in Pseudomonas aeruginosa) doesn’t bind, and then positioned the rhl-box inside an artificial lacZ promoter as illustrated in Fig 4, between and partially overlapping the consensus -35 and -10 hexamers, RNA polymerase binding sites. We also replicated work done by Kristia Egland et al which serves as a positive control of our design.
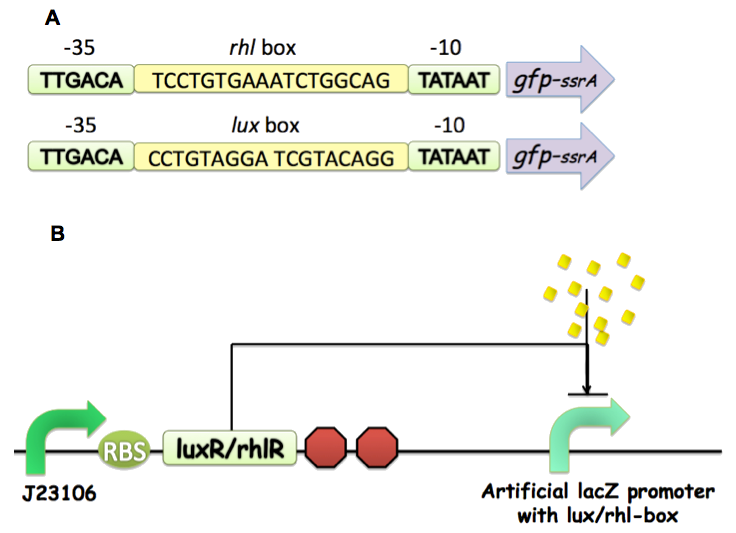
Figure 4: (A) Building Rhl repressible promoter interfaced with appropriate chemical wires. (B) The overall architecture of our quorum sensing repression system.
A GFP reporter with C-terminus fused ssrA degradation tag was used to report the transcriptional activity at promoters. The Rhl repressible promoter-GFP ssrA was then cloned into pSB4A5 backbone and BBa_J23106 (constitutive promoter)-rhlR- BBa_B0015 (terminator) into pSB1K3. The Lux repressible promoter-GFP ssrA was fused with the BBa_J23106 (constitutive promoter)-luxR- BBa_B0015 (terminator) and cloned into pSB1C3.
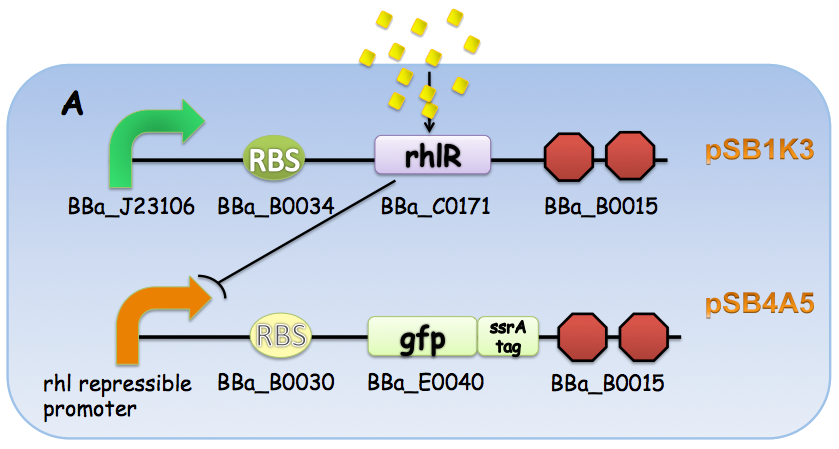
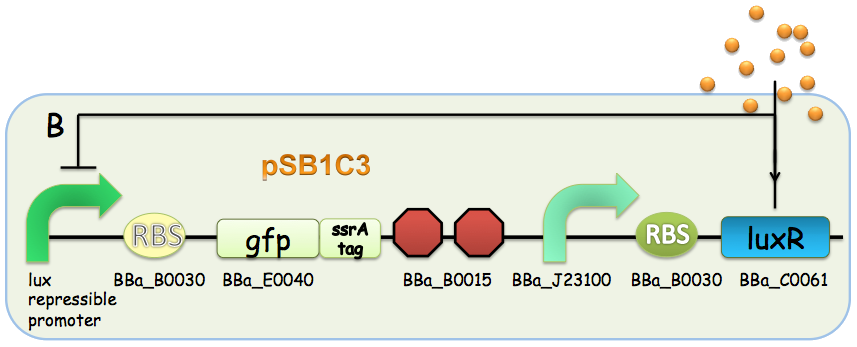
Figure 5: The genetic construction of the circuit design used for characterization. (A) Rhl repressible promoter-rhlR system. (B) Lux repressible promoter-luxR system.
Conversion of transcriptional activators LuxR and RhlR into repressors
To evaluate the properties of lux/rhl repressible promoter, we applied dose response assay and time dependence assay. In these assays GFP intensity was measured to quatitatively evaluate the promoter acitivity by Tecan Microplate Reader with excitation wavelength at 470nm and emission wavelength at 509nm. A black 96-well plate was used to minimize the interference of different well. OD 600 was also measured by Tecan Microplate Reader in a transparent 96-well plate.
Dose response assay
An overnight culture of bacteria grown in LB with antibiotics at 37°C was reactivated by diluting the culture in a ratio of 1:1000 with fresh LB. The LB we used was pre-mixed with different dose of C4-HSL / 3OC6-HSL and its final concentration varied from 0 to 1mM/1μM. When OD600 reached 0.6-0.8, pellet bacterial cells by 4 min centrifugation at 4000 rpm, discard the supernatant. Resuspend the pelleted cells in 500 μl of PBS, and then pippete 200uL of bacterial resuspention into each well of 96-well plate.
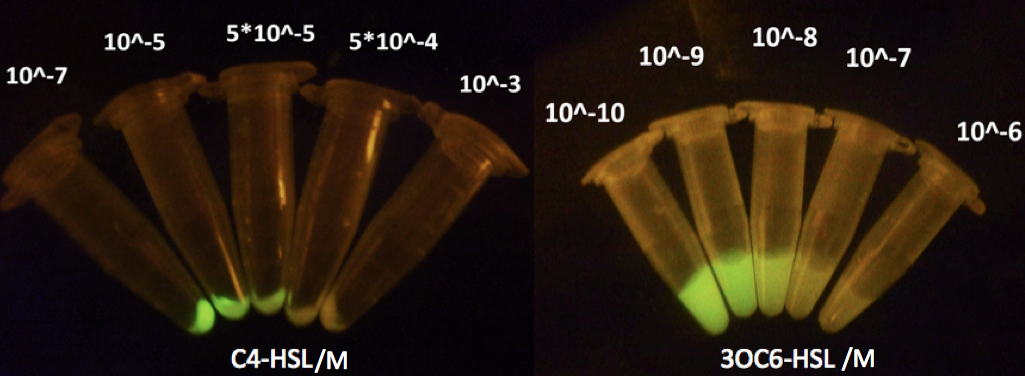
Figure 6: Repression effect of previous transcription activators at artificial AHL repressible promoters under a gradient of C4-HSL / 3OC6-HSL concentrations, respectively.
Figure 6 shows repression of GFP expression in circuits depicted in Fig 5 under gradient concentrations of C4-HSL/3OC6-HSL, which indicates that the activity of Rhl/Lux repressible promoter was C4-HSL/3OC6-HSL dependent, with strongest repression at 5*10^-4 M of C4-HSL and 10nM of 3OC6-HSL.
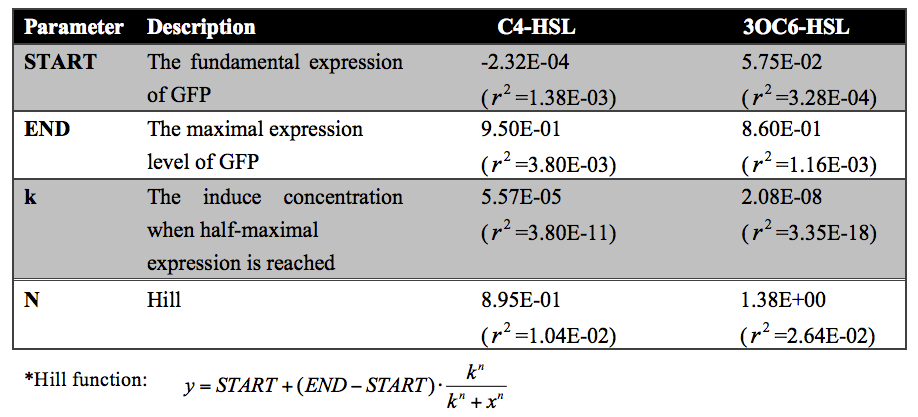
Quantitative analysis was also conducted. As shown in the following figures and tables, both of the dose response curves represent as a hill function and the decrease of transcription activity at promoters was approximate 20-fold.
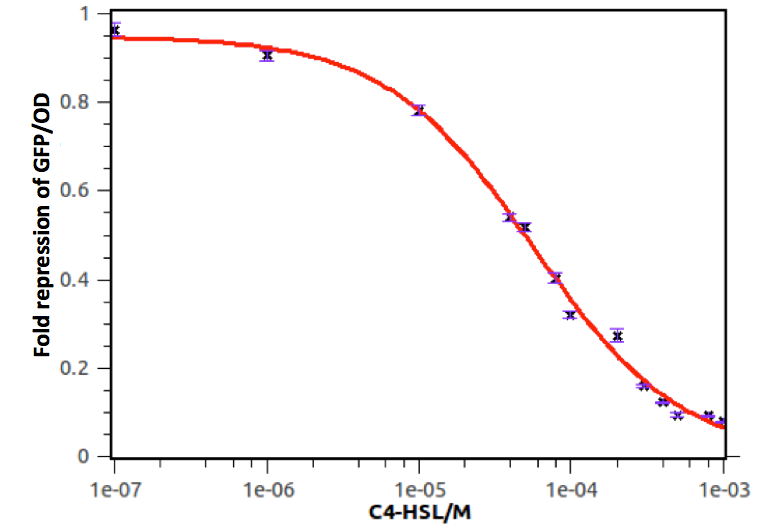
Figure 7: Fold repression of GFP/OD expressed by rhl repressible promoter on C4-HSL signal concentration. In this experiment, GFP expression in E. coli containing pSB1K3 and pSB4A5 was measured at a series of concentrations of C4-HSL. The fold induction of GFP/OD is expressed as the percentage of expression in the absence of C4-HSL. Error bars correspond to the standard deviation from multiple measurements.
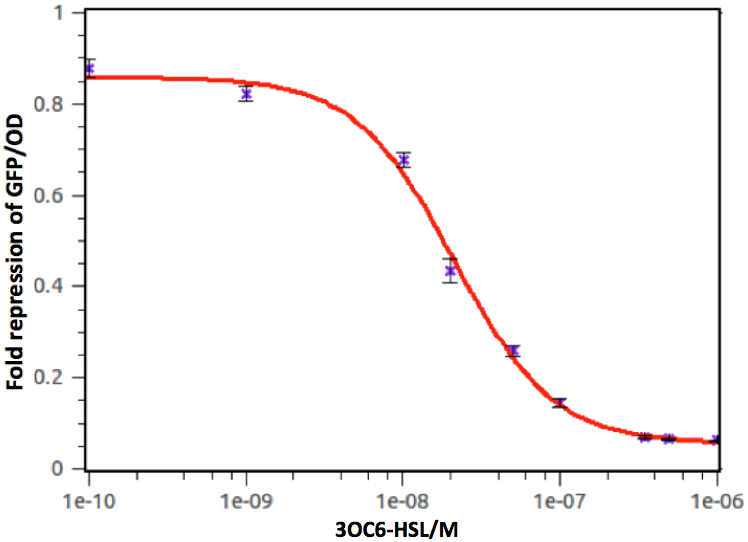
Figure 8: fold repression of GFP/OD expressed by lux repressible promoter on 3OC6-HSL signal concentration. In this experiment, GFP expression in E. coli containing pSB1C3 was measured at a series of concentrations of 3OC6-HSL. The fold induction of GFP/OD is expressed as the percentage of expression in E. coli cells containing pSB1C3 in the absence of 3OC6-HSL. Error bars correspond to the standard deviation from multiple measurements.
Therefore, we have demonstrated that the AHL repressible promoter with rhl/lux-box owes the ability to convert transcriptional activator rhlR/luxR into repressor.
Time dependence assay
An overnight culture of grown in LB with antibiotics at 37°C was reactivated by diluting the culture in a ratio of 1:1000 with fresh LB. When OD600 reached 0.4, the bacteria was disposed to several EP tubes, each owning 500uL, and C4-HSL/3OC6-HSL was supplied with 3 duplicates and the final concentration was 1mM/1μM. We cultured the fluorescence in EP tubes with 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 7, 8 hours at 37°C. Then pellet bacterial cells by 4 min centrifugation at 4000 rpm, discard the supernatant. Resuspend the pelleted cells in 500 μl of PBS, and then pippete 200uL of bacterial resuspention into each well of 96-well plate.
The results of the assay are displayed below.
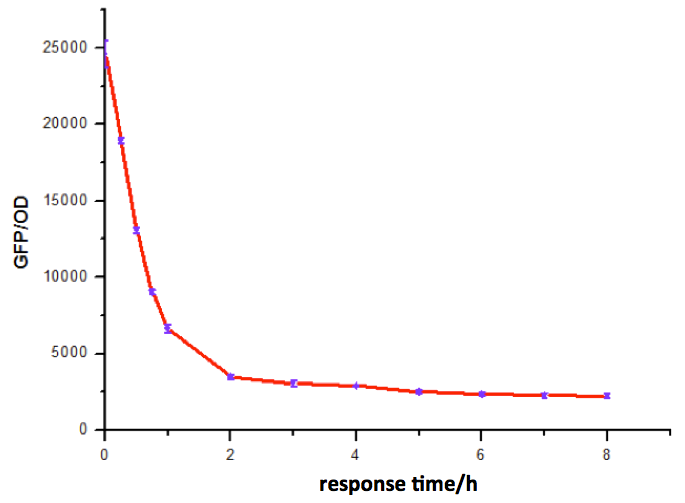
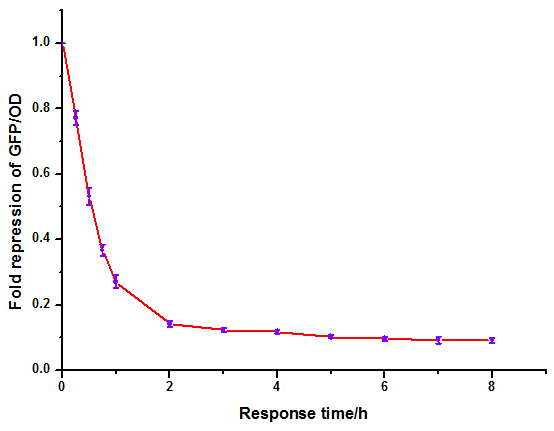
Figure 9: Time dependence of AHL mediated repression at rhl repressible promoter under 1mM of C4-HSL . (A) GFP/OD; (B) fold repression of GFP/OD. Error bars correspond to the standard deviation from multiple measurements.
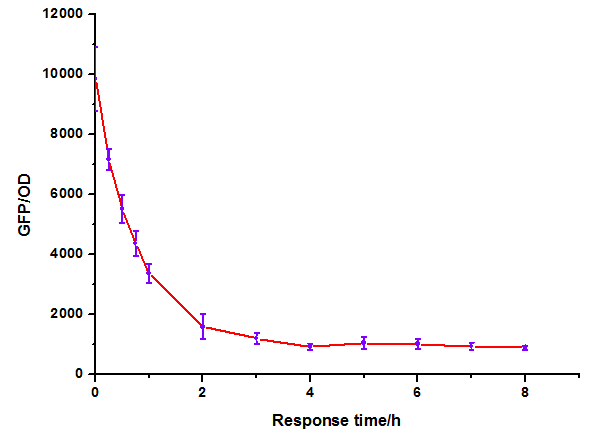
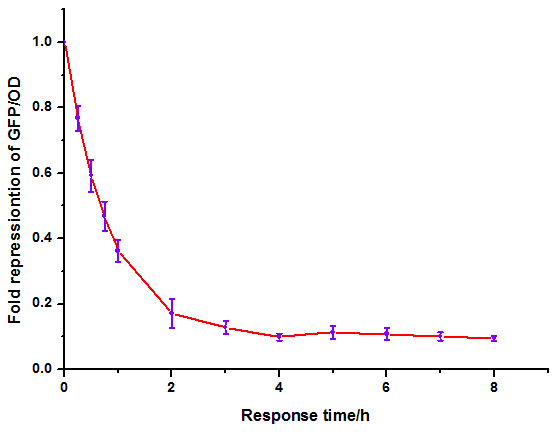
Figure 10: Time dependence of AHL mediated repression at lux repressible promoter under 1mM of C6-HSL. (A) GFP/OD; (B) fold repression of GFP/OD. Error bars correspond to the standard deviation from multiple measurements.
Noting that the GFP expression intensity was reduced from 100% to 10% in merely 2-3 hours, we were delighted by the dramatic repression performance of quorum sensing repressors. The time dependence assay provides further promising clues towards the expansion of our quorum sensing repressor design to more QS systems.
Reference
[1] Li, B. You, L. Synthetic biology: Division of logic labour. Nature 469, 171–172 (2011).
[2] Marguet, P. Balagadde F. Tan C. You L. Biology by design: reduction and synthesis of cellular components and behaviour. JR Soc Interface 4, 607–623 (2007).
[3] Voigt, C.A. Genetic parts to program bacteria. Curr Opin Biotechnol 17, 548–557 (2006).
[4] Clancy, K, Voigt, C.A. Programming cells: towards an automated 'Genetic Compiler'. Curr Opin Biotechnol 21, 572–581 (2010) .
[5] Egland, K.A., Greenberg, E.P. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol 182, 805-811 (2000).
[6] Schuster, M. Urbanowski, M.L. Greenberg, E.P. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. PNAS 45, 15833-15839 (2004).
 "
"