Team:USTC-China/Wet Lab/protocol
From 2011.igem.org
(→Artificial Bacterial barrier Construction and Testing) |
(→Artificial Bacterial barrier Construction and Testing) |
||
| Line 61: | Line 61: | ||
<html><a name="Artificial" id=""></a></html> | <html><a name="Artificial" id=""></a></html> | ||
| - | ==Artificial Bacterial | + | ==Artificial Bacterial Barrier Construction and Testing== |
<p> For the simulation, a cassete containing a ColE7-ImmE7 complex gene([http://partsregistry.org/Part:BBa_K117001 BBa_K117001] ), Toggle Switch-Aptamer-cheZ Device, and Lysis gene([http://partsregistry.org/Part:BBa_K117000 BBa_K117000]) was assembled to create a destruction module. Then using this module to transform the ''E.coli'' RP1616 competence containing the LuxPR-cI Device. After transformation, diluted engineering bacteria and target bacteria suspensions from mid-log-phaase cultures were applied to the sites(as shown in Figure5. ) of the Semi-Solid Media. The plates were dried the room temperature at for 15 min, and incubated at 37°C for 10h to observe the form change of two colonies.Then to further prove our results, we study the relationship between the colE7 released to the media and theophylline gradient by SDS-PAGE.</p> | <p> For the simulation, a cassete containing a ColE7-ImmE7 complex gene([http://partsregistry.org/Part:BBa_K117001 BBa_K117001] ), Toggle Switch-Aptamer-cheZ Device, and Lysis gene([http://partsregistry.org/Part:BBa_K117000 BBa_K117000]) was assembled to create a destruction module. Then using this module to transform the ''E.coli'' RP1616 competence containing the LuxPR-cI Device. After transformation, diluted engineering bacteria and target bacteria suspensions from mid-log-phaase cultures were applied to the sites(as shown in Figure5. ) of the Semi-Solid Media. The plates were dried the room temperature at for 15 min, and incubated at 37°C for 10h to observe the form change of two colonies.Then to further prove our results, we study the relationship between the colE7 released to the media and theophylline gradient by SDS-PAGE.</p> | ||
[[File:C().jpg|center|450px| Figure5.(Colony1: engineered bacteria, Colony2: target bacteria)]] | [[File:C().jpg|center|450px| Figure5.(Colony1: engineered bacteria, Colony2: target bacteria)]] | ||
<p align=center class="ppp">Figure5.(Colony1: engineered bacteria, Colony2: target bacteria)</p> | <p align=center class="ppp">Figure5.(Colony1: engineered bacteria, Colony2: target bacteria)</p> | ||
Revision as of 00:05, 29 October 2011

protocol
1.Aptamer-cheZ Device Construction
2.Dose-Dependent Migration Ability of Aptamer-cheZ Device
3.Toggle Switch Device Testing and Modifying
4.Semi-Solid Media with the Gadient of Theophylline
5.Toggle Switch-Aptamer-cheZ Device Construction and Testing
6.Toggle Switch-Aptamer-cheZ Device(modified) Construction and Testing
7.Artificial Innate Immunity System Construction and Testing
Aptamer-cheZ Device Construction
The cheZ gene was cloned from E.coli TOP10 strain using PCR. A cassete containing a theophylline-sensitive aptamer, and the cheZ gene was assembled using PCR and was subcloned into the SpeI and PstI sites of pSB1A2 containing the lac promoter.
Clone Primer:
Forward:5'-GTTTCGAATTCGCGGCCGCTTCTAGATGCAACCATCAATCAAACC-3'
Reverse:5'-GTTTCCTGCAGCGGCCGCTACTAGTATTATTAAAATCCAAGTCTATCCAACAAATCGT-3'
Assemble Primer:
Forward:5'-GTTTCGAATTCGCGGCCGCTTCTAGGGTGATACCAGCATCGTCTTGATGCCCTTGGCAGCACCCCGCTGCAAGACAACAAG ATGCAACCATCAATCAAACC-3'
Reverse:5'-GTTTCCTGCAGCGGCCGCTACTAGTATTATTAAAATCCAAGACTATCCAACAAATCGT-3'
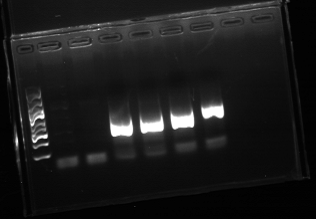
Assemble condition: 96°C 10min, (96°C 30s, 56°C 30s, 72°C 1min)16 cycles and each cycle the annealing temperature increase 1°C, (96°C 30s, 72°C 30s, 72°C 1min)20 cycles,72°C 10min and holding at 4°C.
Figure1.Assemble PCR Result
Dose-Dependent Migration Ability of Aptamer-cheZ Device
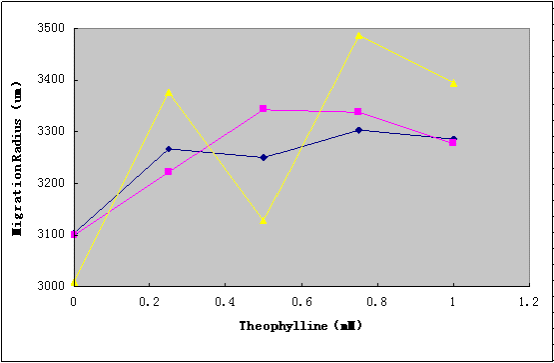
To test the migration ability, selective media (tryptone broth with different ratio of agar(0.25%, 0.3%, 0.4%), 50μg/mL ampicllin, and various concentrations of theophylline(0mM, 0.25mM, 0.5mM, 0.5mM, 0.75mM, 1mM)) was prepared in Petri dishes(85mm dia). Diluted cell suspensions from mid-log-phase cultures were applied to the center of the plates, which were dried at room temperature at for 15 min, and incubated at 37°C for 10h, and the migration radii were determined by measuring the diameter of the outermost ring of growth.
(a)
(b)
(c)
Figure2.((a)0.25%agar, (b)0.3%agar, (c)0.4%agar)
The results(Figure2.) show that the Aptamer-cheZ Device is functional, and the 0.3%agar is the best ratio for the migration experiments.
Toggle Switch Device Testing and Modifying
First, test the function of the original Toggle Switch Device from PKU. After the transformation, the ratio between the colonies with red fluorescent and the colonies with green fluorescent is 8:25.
Then, modify the prototype of the Toggle Switch Device from PKU. The cI gene was subcloned into the SpeI and PstI sites of pSB3t5, a low copy plasmid, containing the LuxPR promoter to create LuxPR-cI Device. After the transformation, prepare the E.coli RP1616 competence containing the LuxPR-cI Device, then use the original Toggle Switch Device to transform this competence.
After the transformation, the ratio between the colonies with red fluorescent and the colonies with green fluorescent is 6:1.
Semi-Solid Media with the Gadient of Theophylline
Media was prepared in 100mm square Petri dishes. Layers(15ml) of selective 0.3% agar containing theophylline(1mM, 0.25mM, 0mM) were poured in the pattern shown in Figure 3.
Figure3.
Each layer would solidify for 50 min before applying the following layer. After all layers were applied, the media should equilibrate at room temperature for 3.0 h.
Toggle Switch-Aptamer-cheZ Device Construction and Testing
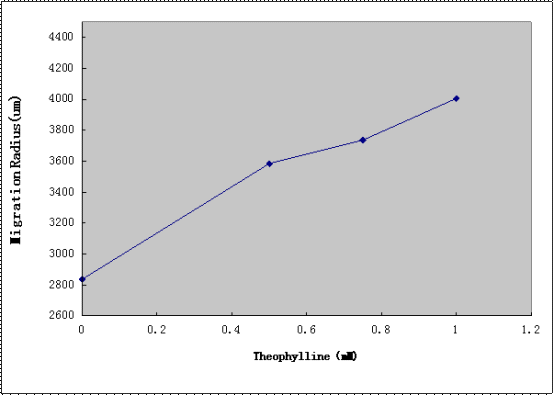
The Aptamer-cheZ Device(without lac promoter) was subcloned into the SpeI and PstI sites of the Toggle Switch Device([http://partsregistry.org/Part:BBa_K228003 BBa_K228003]) from PKU to create the Toggle Switch -Aptamer-cheZ Device. After the transformation, diluted cell suspensions from mid-log-phase cultures were applied to the center of the Semi-Solid Media(as shown in Figure4.). The plates were dried the room temperature at for 15 min, and incubated at 37°C for 10h, and the migration radii were determined by measuring the diameter of the outermost ring of growth.
Figure4.
Toggle Switch-Aptamer-cheZ Device(modified) Construction and Testing
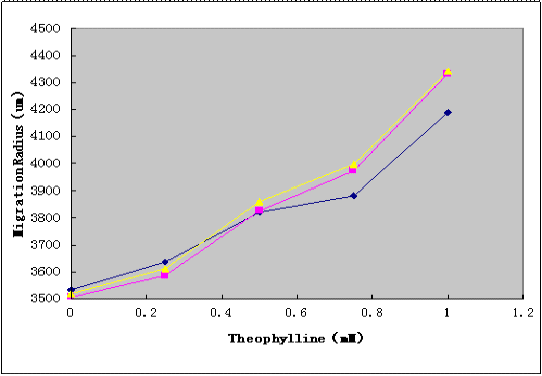
Using Toggle Switch-Aptamer-cheZ Device to transform the E.coli RP1616 competence containing the LuxPR-cI Device. After transformation, diluted cell suspensions from mid-log-phaase cultures were applied to the center of the Semi-Solid Media(as shown in Figure4. ). The plates were dried the room temperature at for 15 min, and incubated at 37°C℃for 10h, and the migration radii were determined by measuring the diameter of the outermost ring of growth.Then to further prove our results, we study the relationship between the cheZ expression and theophylline gradient by SDS-PAGE.
Artificial Bacterial Barrier Construction and Testing
For the simulation, a cassete containing a ColE7-ImmE7 complex gene([http://partsregistry.org/Part:BBa_K117001 BBa_K117001] ), Toggle Switch-Aptamer-cheZ Device, and Lysis gene([http://partsregistry.org/Part:BBa_K117000 BBa_K117000]) was assembled to create a destruction module. Then using this module to transform the E.coli RP1616 competence containing the LuxPR-cI Device. After transformation, diluted engineering bacteria and target bacteria suspensions from mid-log-phaase cultures were applied to the sites(as shown in Figure5. ) of the Semi-Solid Media. The plates were dried the room temperature at for 15 min, and incubated at 37°C for 10h to observe the form change of two colonies.Then to further prove our results, we study the relationship between the colE7 released to the media and theophylline gradient by SDS-PAGE.
Figure5.(Colony1: engineered bacteria, Colony2: target bacteria)
 "
"