Team:USTC-China/Project
From 2011.igem.org
(→Method) |
|||
| Line 21: | Line 21: | ||
<p> So far we have successfully constructed a novel system in which bacterial colonies will automatically divide into two parts after certain time. Over the summer we have been working on assembling [https://2011.igem.org/Team:USTC-China/Project/module#Riboswitch riboswitches] finely tuned by small molecules, which will act as the main power to drive two parts away from each other, and [https://2011.igem.org/Team:USTC-China/Project/module#Toggle-switch toggle switches] pushed on and off while memorizing current state, which will play the role of giving birth to two ‘different’ kinds of bacteria in one colony. Furthermore, we have been modulating the toggle switch to produce a more balanced ratio and creatively integrating [https://2011.igem.org/Team:USTC-China/Project/module#Quorum quorum sensing system] into our system to optimize our [https://2011.igem.org/Team:USTC-China/Project/results results] .</p> | <p> So far we have successfully constructed a novel system in which bacterial colonies will automatically divide into two parts after certain time. Over the summer we have been working on assembling [https://2011.igem.org/Team:USTC-China/Project/module#Riboswitch riboswitches] finely tuned by small molecules, which will act as the main power to drive two parts away from each other, and [https://2011.igem.org/Team:USTC-China/Project/module#Toggle-switch toggle switches] pushed on and off while memorizing current state, which will play the role of giving birth to two ‘different’ kinds of bacteria in one colony. Furthermore, we have been modulating the toggle switch to produce a more balanced ratio and creatively integrating [https://2011.igem.org/Team:USTC-China/Project/module#Quorum quorum sensing system] into our system to optimize our [https://2011.igem.org/Team:USTC-China/Project/results results] .</p> | ||
<p> As to modeling, we have not only been building [https://2011.igem.org/Team:USTC-China/Drylab/modeling models of the movement] but also been collecting and analyzing data for an [https://2011.igem.org/Team:USTC-China/Drylab/riboswitch aptamers analysis] for small molecules and corresponding genomic sequences and structures in guiding bacteria.</p> | <p> As to modeling, we have not only been building [https://2011.igem.org/Team:USTC-China/Drylab/modeling models of the movement] but also been collecting and analyzing data for an [https://2011.igem.org/Team:USTC-China/Drylab/riboswitch aptamers analysis] for small molecules and corresponding genomic sequences and structures in guiding bacteria.</p> | ||
| - | <p> Finally, we are intended to construct a [https://2011.igem.org/Team:USTC-China/Project/application Artificial Bacterial Barrier] with our self-organized bacteria, in which | + | <p> Finally, we are intended to construct a [https://2011.igem.org/Team:USTC-China/Project/application Artificial Bacterial Barrier] with our self-organized bacteria, in which engineered intestinal bacteria is used as a safety weapon to destroy the pathogens which invade the intestinal mucosal system.</p> |
<html><a name="Introduction" id=""></a></html> | <html><a name="Introduction" id=""></a></html> | ||
| Line 30: | Line 30: | ||
<p> Now the key problem is to produce two different kinds of bacterial in one colony, in which one kind could be guided by riboswitches mentioned above while the other would keep still or move to the opposite side. The work of PKU-IGEM 2007 provides us with a ideal platform in which bacterial are equipped with devices to differentiate out of homogeneous conditions.Thanks to their warm help, we acquire Toggle-switches and made efforts to modulate the switch into to produce a more balanced ratio, so that the results would be more likely to be apparently visible.</p> | <p> Now the key problem is to produce two different kinds of bacterial in one colony, in which one kind could be guided by riboswitches mentioned above while the other would keep still or move to the opposite side. The work of PKU-IGEM 2007 provides us with a ideal platform in which bacterial are equipped with devices to differentiate out of homogeneous conditions.Thanks to their warm help, we acquire Toggle-switches and made efforts to modulate the switch into to produce a more balanced ratio, so that the results would be more likely to be apparently visible.</p> | ||
<p> With the help of modeling, it’s spotted that the part driven away cannot be efficiently turned into another colony compared with the other part staying, since bacteria tend to move instead of grouping on their way towards the most concentrated location of the chemical signal. Quorum sensing, as another most applied mechanism in synthetic biology, give us a hint in solving this problem.</p> | <p> With the help of modeling, it’s spotted that the part driven away cannot be efficiently turned into another colony compared with the other part staying, since bacteria tend to move instead of grouping on their way towards the most concentrated location of the chemical signal. Quorum sensing, as another most applied mechanism in synthetic biology, give us a hint in solving this problem.</p> | ||
| - | <p> The final step is to picture the blueprint of the application of our system. Here we present one in medical area, in which | + | <p> The final step is to picture the blueprint of the application of our system. Here we present one in medical area, in which engineered intestinal bacteria can help you with pathogens invasion. (Artificial Innate Immunity System) Given the difficulties of human experiment, we conduct a simulation in E.coli.</p> |
<html><a name="Methods" id=""></a></html> | <html><a name="Methods" id=""></a></html> | ||
| Line 56: | Line 56: | ||
<html> | <html> | ||
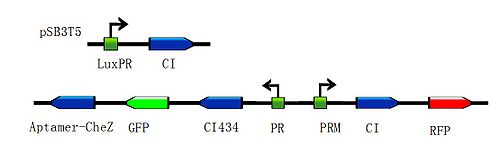
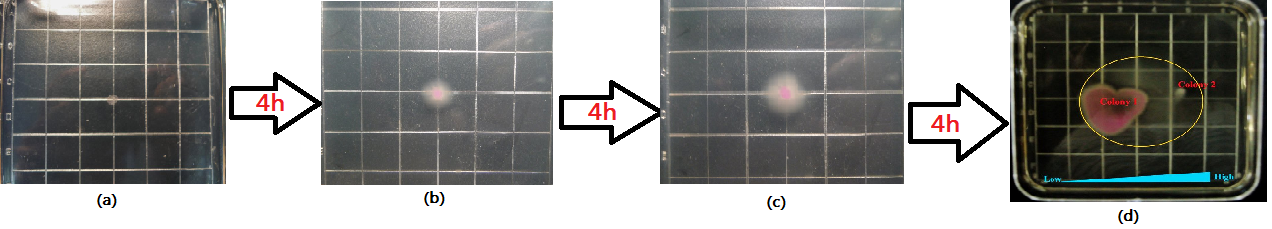
| - | <div class="center"><div class="thumb tnone" style="width:602px;"><div class="thumbinner" style="width:602px;"><a href="/File:G().jpg" class="image"><img alt="G().jpg" src="/wiki/images/thumb/e/ee/G%28%29.jpg/600px-G%28%29.jpg" width="600" height="183" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:G().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></font><font size="2" align="center">Figure 3:According to the basic design and we assume the small molecule is theophylline, so under the control of this module, the | + | <div class="center"><div class="thumb tnone" style="width:602px;"><div class="thumbinner" style="width:602px;"><a href="/File:G().jpg" class="image"><img alt="G().jpg" src="/wiki/images/thumb/e/ee/G%28%29.jpg/600px-G%28%29.jpg" width="600" height="183" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:G().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></font><font size="2" align="center">Figure 3:According to the basic design and we assume the small molecule is theophylline, so under the control of this module, the engineered bacteria with green fluorescence will move toward the infection site with a high concentration of theophylline and the expression of Lysis becomes more and more, while the engineered bacteria with red fluorescence will stay at the original site and wait to creat the second wave of the engineered bacteria with green fluorescence, they have been protected from being destroyed by the pyosin from the engineered bacteria with green fluorescence. The engineered bacteria with green fluorescence will destroy themselves and release the Pyosin, which is expressed at the beginning of the process, to kill the pathogens.(Images by USTC-IGEM with TinkerCell)</font><font size="3"></div></div></div></div> |
</html> | </html> | ||
| Line 72: | Line 72: | ||
<p align=center class="ppp"></p> | <p align=center class="ppp"></p> | ||
| - | <p> Figure5 clearly display the dividing line between the | + | <p> Figure5 clearly display the dividing line between the engineered bacteria at two different states, and the bacteria mainly express GFP indeed moving towards the high concentration of theophylline. This means the modified Toggle-switch-Aptamer-cheZ actually work as our design.</p> |
Revision as of 23:29, 28 October 2011

Contents |
Overview
1.Abstract
2.Introduction/motivation
3.Methods
4.Results and discussion
5.Reference
Abstract
So far we have successfully constructed a novel system in which bacterial colonies will automatically divide into two parts after certain time. Over the summer we have been working on assembling riboswitches finely tuned by small molecules, which will act as the main power to drive two parts away from each other, and toggle switches pushed on and off while memorizing current state, which will play the role of giving birth to two ‘different’ kinds of bacteria in one colony. Furthermore, we have been modulating the toggle switch to produce a more balanced ratio and creatively integrating quorum sensing system into our system to optimize our results .
As to modeling, we have not only been building models of the movement but also been collecting and analyzing data for an aptamers analysis for small molecules and corresponding genomic sequences and structures in guiding bacteria.
Finally, we are intended to construct a Artificial Bacterial Barrier with our self-organized bacteria, in which engineered intestinal bacteria is used as a safety weapon to destroy the pathogens which invade the intestinal mucosal system.
Introduction/Motivation
Cells grow and cells divide. This is the basis of life and nature. In our eyes, a bacterial colony, which is also formed by bacterial growth and division, is a big cell too. Given that bacteria always play their roles as a ‘group’ instead of an ‘individual’, a bacterial colony can be considered as the unit of our intended function. It may not be hard to imagine if this unit could grow and divide itself automatically, significant applications would be developed. In fact, similar ideas have already been realized in eukaryotic systems.
Chemotaxis has always been a powerful tool in organizing bacteria’s movement and it’s always an important ability to reprogram bacteria to follow entirely new chemical signals. Riboswitches, which have been a hot issue of synthetic biology recently, can be used to guide E.Coli toward and precisely localized to a completely new chemical signal. We take advantage of them in our system to act as the main power to drive two parts of a bacterial colony away from each other. Here we really want to thank Professor J. S. Parkinson for providing ΔcheZ strain RP1616 and the corresponding wild type RP437 and Professor J. P. Gallivan for directing our construction of riboswitches.
Now the key problem is to produce two different kinds of bacterial in one colony, in which one kind could be guided by riboswitches mentioned above while the other would keep still or move to the opposite side. The work of PKU-IGEM 2007 provides us with a ideal platform in which bacterial are equipped with devices to differentiate out of homogeneous conditions.Thanks to their warm help, we acquire Toggle-switches and made efforts to modulate the switch into to produce a more balanced ratio, so that the results would be more likely to be apparently visible.
With the help of modeling, it’s spotted that the part driven away cannot be efficiently turned into another colony compared with the other part staying, since bacteria tend to move instead of grouping on their way towards the most concentrated location of the chemical signal. Quorum sensing, as another most applied mechanism in synthetic biology, give us a hint in solving this problem.
The final step is to picture the blueprint of the application of our system. Here we present one in medical area, in which engineered intestinal bacteria can help you with pathogens invasion. (Artificial Innate Immunity System) Given the difficulties of human experiment, we conduct a simulation in E.coli.
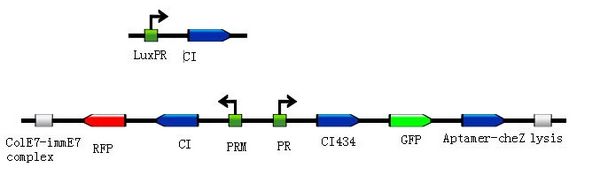
Method
- Verification of ΔcheZ strain
- Construction of Riboswitch: Theophylline guided riboswitch + cheZ
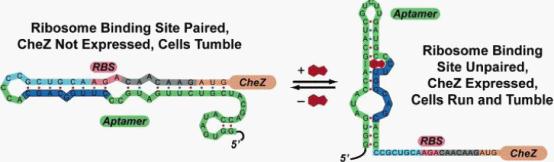
In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811)
- Verification of Toggle-switch
- Modulate Toggle-switches to produce more balanced ratio
- Incorporation of Riboswitch into one side of Toggle-switch
- Verification of the constructed system’s function
- Application: simulation of Artificial Bacterial Barrier
Results and Discussions
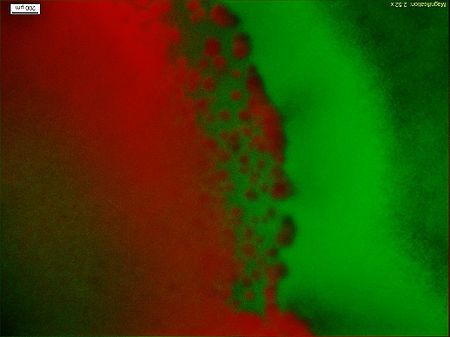
Figure5 clearly display the dividing line between the engineered bacteria at two different states, and the bacteria mainly express GFP indeed moving towards the high concentration of theophylline. This means the modified Toggle-switch-Aptamer-cheZ actually work as our design.
References
- 1.Timothy S. Gardner, Charles R. Cantor, James J. Collins (2000) Construction of a Genetic toggle switch in Escherichia coli. Nature, 403,339-342
- 2.Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811
- 3.Chunbo Lou, Xili Liu, Ming Ni.etc (2010) Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Molecular Systems Biology, 6:350
- 4.Joy Sinha, Samuel J Reyes, Justin P Gallivan (2010) Reprogramming bacteria to seek and destroy an herbicide. Nature Chemical Biology, 6,464-470
- 5.Steven L. Porter, George H. Wadhams, Judith P. Armitage (2011) Signal processing in complex chemotaxis pathways. Nature Reviews Microbiology, 9,153-165
- 6.Warren C. Ruder, Ting Lu, James J. Collins (2011) Synthetic Biology Moving into the Clinic. Science,333,1248-1252
- 7.Nazanin Saeidi, Choon Kit Wong, Tat-Ming Lo .etc (2011) Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology,7:251
 "
"