Team:NYMU-Taipei/results/immunological-solution1
From 2011.igem.org
(→Symbiosis with Human Glial Cell) |
(→Symbiosis with Human Glial Cell) |
||
| Line 19: | Line 19: | ||
==<font size=5><font color=crimson>'''Symbiosis with Human Glial Cell '''</font></font>== | ==<font size=5><font color=crimson>'''Symbiosis with Human Glial Cell '''</font></font>== | ||
| - | <font size=3>It is well-studied in <i>Magnetotactic bacteria were introduced into guanulocytes and monocytes by phagocytosis.</i>(Tadashi Matsunaga et all. 1986) In order to simulate the circumstances in brain, we put <i>E.coli</i> and <i>AMB-1</i> separately into mixed glial cells. Below are the photos we captured under microscope.</font> | + | <font size=3>It is well-studied in <i>Magnetotactic bacteria were introduced into guanulocytes and monocytes by phagocytosis.</i>(Tadashi Matsunaga et all. 1986) In order to simulate the circumstances in brain, we put <i>E.coli</i> and <i>AMB-1</i> separately into mixed glial cells <i>in vitro</i>. Below are the photos we captured under microscope.</font> |
==<font size=5><font color=crimson>'''Measurements of Construct <i>tetR</i>+<i>rbs</i>+<i>gfp</i> under Fluorescence Microscope'''</font></font>== | ==<font size=5><font color=crimson>'''Measurements of Construct <i>tetR</i>+<i>rbs</i>+<i>gfp</i> under Fluorescence Microscope'''</font></font>== | ||
Revision as of 02:54, 6 October 2011

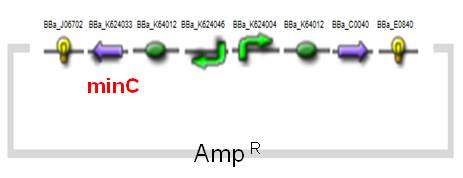
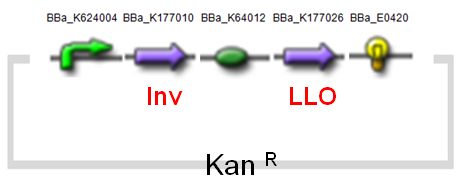
Construction & Parts
Our minC construct and Inv & LLO construct
| | |
Symbiosis with Human Glial Cell
It is well-studied in Magnetotactic bacteria were introduced into guanulocytes and monocytes by phagocytosis.(Tadashi Matsunaga et all. 1986) In order to simulate the circumstances in brain, we put E.coli and AMB-1 separately into mixed glial cells in vitro. Below are the photos we captured under microscope.
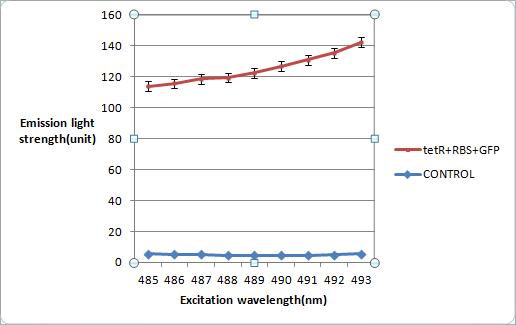
Measurements of Construct tetR+rbs+gfp under Fluorescence Microscope
| 
| 
|
|---|
These are the measurements of our construct [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624001 tetR+rbs+gfp](BBa_K624001) under fluorescence microscope. The left graph is from the control group of E. coli strain DH5-alpha. The middle is tetR+rbs+gfp in plasmid pSB1C3 in E. coli, while the right is from a plate without any bacilli. All graphs are measured in fixed exposure time. Construct of tetR+rbs+gfp which emits light is thought to be a leak phenomenon of plasmid, while RE digest has proved that the sequence length is correct, and further sequencing work is under operation.
gfp measurements under different excitation wavelengths
The expression of gfp is measured precisely under excitation wavelength from 475nm to 501nm, given the absorption wavelength 511nm. In our measurement, wavelength over 493nm yields high background, resulting in values of control group surging far beyond normal condition. Thus, 493nm is suggested as the best excitation wavelength, as it presents the highest emission light and the lowest of the control group.
 "
"