Team:NYMU-Taipei/chassis
From 2011.igem.org
Cheyennexym (Talk | contribs) (→Related Parts) |
Cheyennexym (Talk | contribs) (→Background and Origin) |
||
| Line 15: | Line 15: | ||
---- | ---- | ||
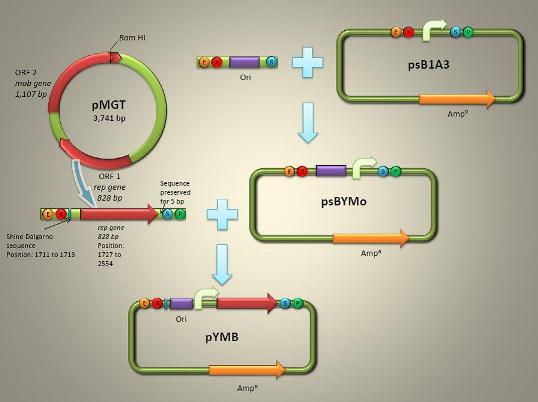
| - | <font size=3>In order to create a shuttle vector for manipulating and transforming gene into Magnetospirillum magneticum AMB-1, we found sound theoretical supports from papers published from Matsunaga T.’s laboratory in Tokyo University of Agriculture and Technology. Under their research and information support, we know that the construct of pUMG is very simple, with pUC19 ligated on BamHI site with pMGT, an endogenous plasmid found in Magnetospirillum magneticum MGT-1. | + | <font size=3> In order to create a shuttle vector for manipulating and transforming gene into Magnetospirillum magneticum AMB-1, we found sound theoretical supports from papers published from Matsunaga T.’s laboratory in Tokyo University of Agriculture and Technology. Under their research and information support, we know that the construct of pUMG is very simple, with pUC19 ligated on BamHI site with pMGT, an endogenous plasmid found in Magnetospirillum magneticum MGT-1. |
Therefore, the construction of a new shuttle vector named pYMB is our primary aim of our cooperative initiative.</font> | Therefore, the construction of a new shuttle vector named pYMB is our primary aim of our cooperative initiative.</font> | ||
Revision as of 02:33, 6 October 2011

Contents |
Plasmid Backbones We name it pYMB...
Background and Origin
In order to create a shuttle vector for manipulating and transforming gene into Magnetospirillum magneticum AMB-1, we found sound theoretical supports from papers published from Matsunaga T.’s laboratory in Tokyo University of Agriculture and Technology. Under their research and information support, we know that the construct of pUMG is very simple, with pUC19 ligated on BamHI site with pMGT, an endogenous plasmid found in Magnetospirillum magneticum MGT-1. Therefore, the construction of a new shuttle vector named pYMB is our primary aim of our cooperative initiative.
BLOC MAGNETO Worldwide Forum
Structure and Assembly
pYMB(Fig. 1) is described to contain the ori (origin of replication), rep gene (required for replication) of pMGT. Once the synthetic work had been done, pYMB is constructed by equipping the ori, the appropriate promoter for AMB-1 (Pmms16 and Pmsp3 as our candidate) and rep gene on the commercial plasmid pUG19 which was revised as the expression of Biobrick backbone psB1A1with promotor Pmsp1. The constructed vector is capable of replicating within both E. coli and AMB-1, fully sufficing a competent shuttle vector for genetic engineering the magnetotactic bateria.
Related Parts
We have created several PARTs on the basis of Biobrick. The expression will be shown on the partregistry webpage.
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624004 BBa_K624004] | ori+Pmsp1+rep (pYMB essentials) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624017 BBa_K624017] | pYMB essentials+(Y-13-R)+(CHAMP) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624018 BBa_K624018] | pYMB essentials+(YN-13-R)+(YC-CHAMP) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624019 BBa_K624019] | pYMB essentials+(13-R)+(Y-CHAMP) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624020 BBa_K624020] | pYMB essentials+(13-R) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624021 BBa_K624021] | pYMBG (rbs: Pmsp3) essentials |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624022 BBa_K624022] | pYMBG (rbs: trunc. Pmsp3) essentials |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624023 BBa_K624023] | pYMB essentials+(Y-13-R) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624024 BBa_K624024] | pYMB essentials+(YN-13-R) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624028 BBa_K624028] | pYMB+rbs (Pmsp3) (expression vector essentials on Magnetospirillum magneticum AMB-1) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624029 BBa_K624029] | pYMB+rbs (Pmsp3, 6 bps trunc.) (expression vector essentials on Magnetospirillum magneticum AMB-1) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624030 BBa_K624030] | pYMB+(13-R)+(CHAMP-Y) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624031 BBa_K624031] | pYMB essentials+(YN-13-R)+(CHAMP-YC) |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624044 BBa_K624044] | pYMB essentials + RBS + LLO + RBS + Invasin + RBS + ECFP |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624051 BBa_K624051] | pYMB essentials + RBS(trunc.) + LLO + RBS(trunc.) + Invasin + RBS(trunc.) + ECFP |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624055 BBa_K624055] | pYMB essentials + RBS(Pmsp3) + tetR + RBS(Pmsp3) + GFP |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624056 BBa_K624056] | pYMB essentials + RBS(Pmsp3) + minC |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624057 BBa_K624057] | pYMB essentials + RBS(Pmsp3) + LLO |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624058 BBa_K624058] | pYMB essentials + RBS(Pmsp3) + Inv |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624060 BBa_K624060] | pYMB essentials + RBS(trunc.) + minC |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624061 BBa_K624061] | pYMB essentials + RBS(trunc.) + LLO |
| [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624062 BBa_K624062] | pYMB essentials + RBS(trunc.) + Inv |
Promoters
Constitutive
Negatively Regulated
Ribosomal Binding Sites
Cultivation of Magnetospirillum magneticum AMB-1 Actually, it was not difficult...
Like E. coli, AMB-1 also grows in liquid medium, and this medium is called Magnetic Spirillum Growth Medium (MSGM). In the medium, we added nutrient, iron source (supplying AMB-1 in magnetite formation within magnetosomes), vitamin, and mineral solution.
MSGM's ingredients
(In grams per liter)
●Succinic acid 0.74 g ●KH2PO4 0.68 g ●Sodium nitrate 0.12 g ●Sodium thioglycolate 0.1 g ●*Wolfe's vitamin solution 10 ml(stock) ●*Wolfe’s mineral solution 5 ml(stock) ●Ferric quinate solution 2 ml(0.27 g of FeCl3 and 0.19 g of quinic acid in 100 ml water) Adjust pH to 7.0 with NaOH
We also refered to an [http://www.sciencedirect.com/science/article/pii/S014102290100343X original article] published by Chen-Dong Yang, et al., which added Polypeptone and Yeast extract into MSGM and added L-cysteine instead of sodium thioglycolate, a mixture called En-rich MSGM. In <<Effects of growth medium composition, iron sources and atmospheric oxygen concentrations on production of luciferase-bacterial magnetic particle complex by a recombinant Magnetospirillum magneticum AMB-1,2001>> said that MSGM enriched with L-cysteine, yeast extract and polypeptone could enhance BMP (Bacterial Magnetic Particles) productivity. Addition of yeast extract had no effect on BMP (Bacterial Magnetic Particles) production and polypeptone only improved the final cell density and therefore slightly improved BMP (Bacterial Magnetic Particles) production, whereas L-cysteine induced cell growth!
En-rich MSGM's ingredients
(In grams per liter)
●Succinic acid 0.74 g ●KH2PO4 0.68 g ●Sodium nitrate 0.12 g ●*Wolfe's vitamin solution 10 ml(stock) ●*Wolfe’s mineral solution 5 ml(stock) ●Ferric quinate solution 2 ml(0.27 g of FeCl3 and 0.19 g of quinic acid in 100 ml water) ●L-cysteine 0.005%(Instead of sodium thioglycolate) ●Yeast extract 0.01% ●Polypeptone 0.02% Adjust pH to 7.0 with NaOH
Strict anaerobic conditions have been thought to facilitate BMP (Bacterial Magnetic Particles) production in AMB-1 cells. To induce AMB-1 forming magnetosome, we have to control low O2 concentration.
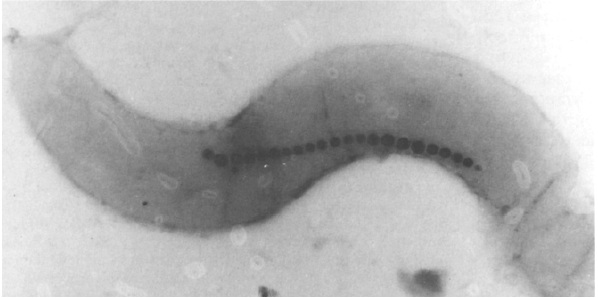
Electroporation So...how to insert our plasmid into the AMB-1?
After plasmid constructions were done, we need to transform them into AMB-1. Electroporation was performed with a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.), at a capacitance of 25 micro-F and a resistance of 200Ω, and 0.1-cm cuvettes. Electroporation allows cellular introduction of large highly charged molecules such as DNA which would never passively diffuse across the hydrophobic bilayer core. This phenomenon indicates that the mechanism is the creation of nm-scale water-filled holes in the membrane.
Procedure:
● Step1: Harvested and washed AMB-1 with 10 mM TES buffer containing 272 mM sucrose (pH 7.5). ● Step2: Resuspended AMB-1 in the same buffer at 10^9 cells/ml. ● Step3: Transferred to 500 micro-liter of MSGM supplemented with 20 mM Mg2+ and incubated at 27°C overnight with shaking at 100 rpm. ● Step4: Diluted in 5 ml of MSGM containing 0.7% agar. ● Step5: Plated on 1% agar in MSGM (supplied with ampicillin at 5 ug/ml or kanamycin at 2.5 ug/ml) incubated under anaerobic conditions.
 "
"