Team:NYMU-Taipei/results/optomagnetic-design1
From 2011.igem.org
(Difference between revisions)
CHIACHENHSU (Talk | contribs) (→The Beginning) |
CHIACHENHSU (Talk | contribs) (→The Beginning) |
||
| Line 21: | Line 21: | ||
The correcterized gel electrophoresis result is shown below. The detail of Mms13's information, please link [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624005]. | The correcterized gel electrophoresis result is shown below. The detail of Mms13's information, please link [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624005]. | ||
| + | |||
| + | [[Image:13_NYMU.png|frame|none|Fig. 2:The correcterized gel electrophoresis result of Mms13.]] | ||
Then, after we have the fundamental material of Mms13, we did the next step of our construct.(See Figure 2) | Then, after we have the fundamental material of Mms13, we did the next step of our construct.(See Figure 2) | ||
Revision as of 02:32, 6 October 2011

Six Constructs and Experimental Results
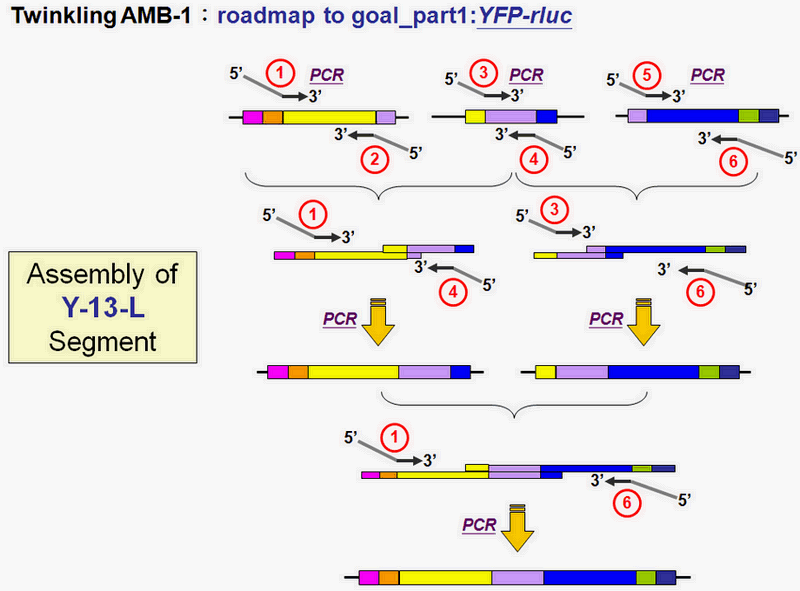
We now follow the steps we construct our design to examine and illustrate what we have done so far. The total parts we construct and their detailed information will be performed and recorded in our part registry.
The Beginning
As soon as we have the AMB-1 bacteria colony, we do the AMB-1 colony PCR to get the Mms13's DNA sequences as our parts to do the following steps.
The correcterized gel electrophoresis result is shown below. The detail of Mms13's information, please link [http://partsregistry.org/wiki/index.php?title=Part:BBa_K624005].
Then, after we have the fundamental material of Mms13, we did the next step of our construct.(See Figure 2)
, The correct and check

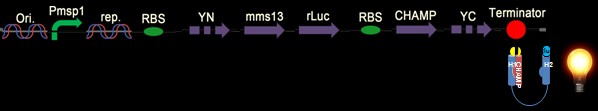
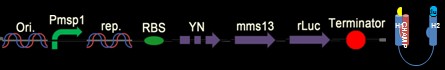
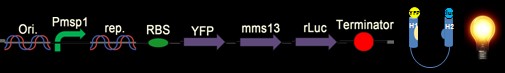
Fig. 7: The design construct is similar with construct in Fig. 5. Why we construct this design with the whole YFP on the C-terminus of the CHAMP design instead of Mms13’s N-terminus is that we try to avoid the interferences of DNA transformation with sequences added in front of the natural construct. The construct is supposed to light up when force pull the Mms13 and turn off if no magnetic field is applied.
 "
"