Team:Kyoto/Digestion
From 2011.igem.org
(→1. Introduction) |
(→1. Introduction) |
||
| Line 8: | Line 8: | ||
| - | ''Streptomyces'' is a kind of prokaryotic bacteria and they produce chitinase. The gene of chitinase had been cloned from ''S. avermitilis'' and introduced into ''Escherichia coli''. | + | ''Streptomyces'' is a kind of prokaryotic bacteria and they produce chitinase. The gene of chitinase had been cloned from ''S. avermitilis'' and introduced into ''Escherichia coli''. Additionally, since the secretion-signal sequences are included in this gene, the chitinase goes out without occurring cell lysis.<br> |
Revision as of 01:01, 6 October 2011
Contents |
Project Digestion
1. Introduction
An insect body is covered with a hard integument containing mainly chitin. To decompose the integument, we used ChiA gene, which encodes chitinase. In order to measure the chitinase activity of the culture supernatant, we evaluated the effects of the medium and cell growth.
Streptomyces is a kind of prokaryotic bacteria and they produce chitinase. The gene of chitinase had been cloned from S. avermitilis and introduced into Escherichia coli. Additionally, since the secretion-signal sequences are included in this gene, the chitinase goes out without occurring cell lysis.
For characterizing the chitinase activity, we used the DNS assay, which is the method to determine the quantity of reducing sugar. In our assay, we determine the quantity of N-acetylglucosamine in a culture supernatant. Since the other materials in the supernatant can react with DNS, it was needed to examine if this assay is available for the quantitative characterization of chitinase activity. We also made a model to assess the enzyme activity in the supernatant from the pre-experiment.
2. Method
Construction
We created following construction to allow secretion of chitinase, chiA1. This gene is regulated by strong lactose promoter, BBa_R0011. We used Streptmyces’s RBS into this constructions, because reference article [1] used that to allow E.coli to secrete the protein.
要変更→
Assay
We performed 3,5-Dinitrosalicylic acid assay (DNS assay), because this assay takes a little time, costs a little money and was used in the previous article measuring chitinase activities [1]. DNS assay is based on this fact: 3,5-dinitorosalicylic acid (DNS), whose color is yellow, reacts with reducing saccharide by boiling and changes into 3-amino-5-nitorosalicylic acid, whose color is brawn.
The more the amount of reducing sugar is, the more this coloring reaction proceeds. We can quantitatively evaluate the degree of this coloring reaction by measuring OD 550, because OD 550 is in direct proportion to the amount of reducing sugar.
We will react DNS reagent with the media where chitin and E.coli introduced chitinase gene are added. If chitinase is secreted in media, chitin is decomposed into reducing sugar, for example, N-acetylglucosamin and coloring reaction proceeds. Therefore, by measuring OD 550, we can indirectly measure chitinase activities.
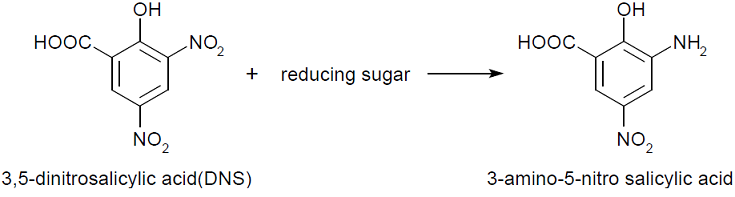
3. Result
This figure shows the overall of DNS assay
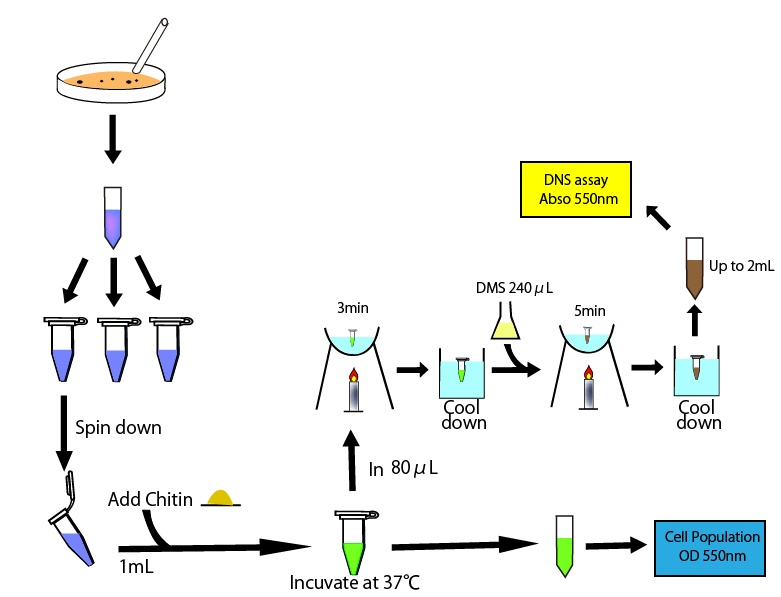
To measure chitinase activity by DNS assay, we thought to need four following things to get accurate result. We need
- to plot the relationship of reducing sugar concentration and OD 550
- to evaluate the impact of components of media on OD 550
- to evaluate the impact of remeineded E.coli in media on OD 550
- to measure chitinase activity
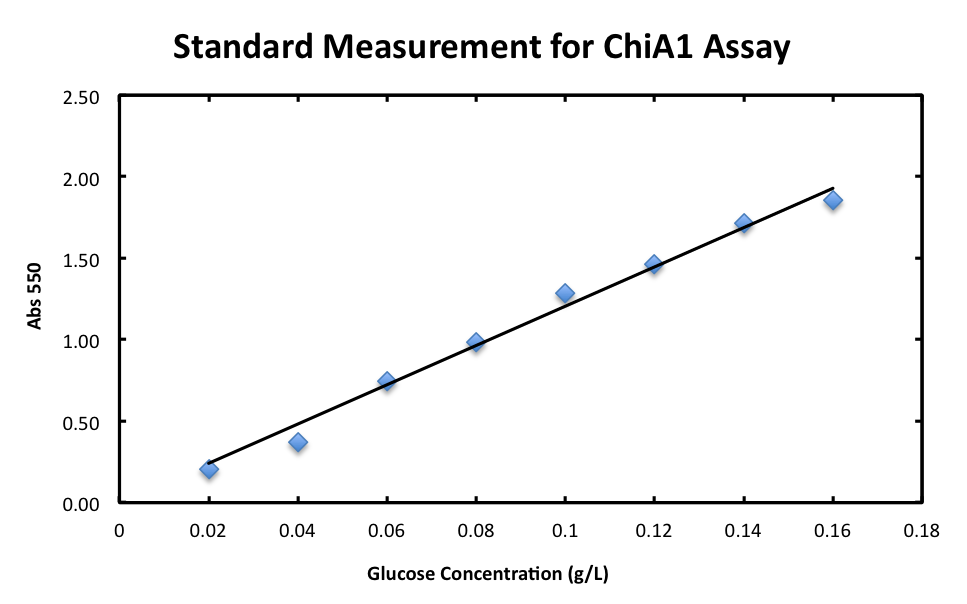
3-1. Standard Measurement for ChiA1.
In order to measure the product of the enzyme reaction, N-acetylglucosamin, quantitatively, we prepared the standard curve using glucose as standard. It is known that the correlation between glucose concentration and the absorbance can be plotted in linaer.
From the result, a strong correlation between glucose concentration and its A550 was observed.
3-2. Consideration of medium and growth of E.coli.
To get accurate values of absorbance, it is needed to consider the remained cells in supernatant. Since we incubate the supernatant for hours for the enzyme reaction, cells can grow, produce chitinase and consume glucose, N-acetylglucosamin, etc; this can influence to DNS assay.
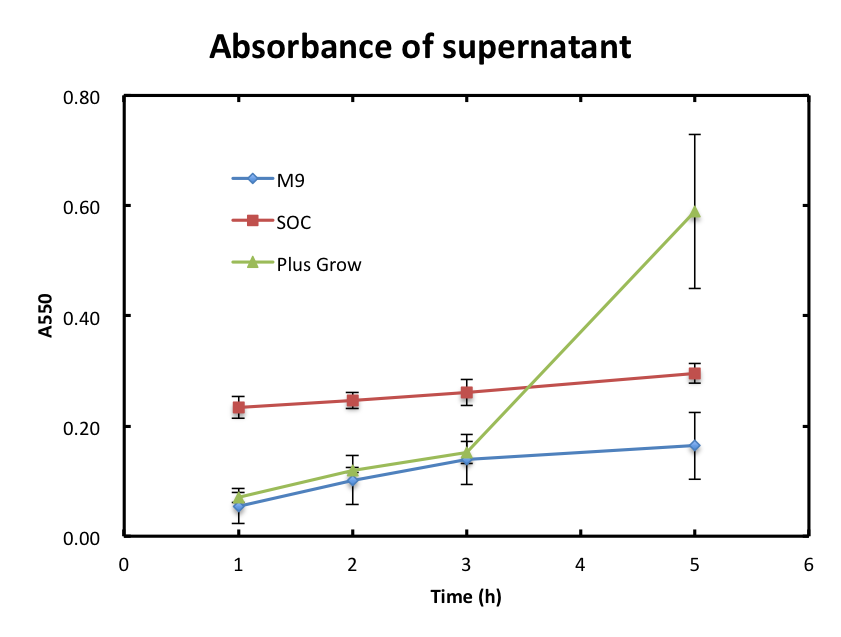
- 3-2-1 The absorbance of supernatant itself.
- In this experiment, we checked if cells were still observed in the supernatant and assayed the growth behavior of cells. Figure 2 shows the A550 of supernatant with time. The absorbance values of M9 were increased slowly and they were smaller than 0.2 during the incubation. The absorbance values of SOC were also increased slowly and they were higher than those of M9. The absorbance of values of Plusgrow Ⅱ were increased obviously between the 3h and 5h.
- 3-2-2 The DNS assay of supernatant.
- From figure 2, the increase in absorbance of Plusgrow Ⅱ was thought to be due to the cell growth. So, we considered that cells still existed in M9 and SOC medium though the significant cell growth couldn't observed.
- In this experiment, we evaluated the influence of cells and its growth to the DNS assay. In other words, we confirmed if cells consume glucose and affect the DNS assay. Figure 3 shows the absorbance of each culture supernatant. The values of M9 and Plusgrow Ⅱ were almost zero during the incubation. The absorbance of SOC was observed, but there was little change during the incubation.
From figure 2 and 3, the cell growth didn't affect to the absorbance of supernatant. About M9 and SOC medium, each medium contains only glucose as sugar source and it is considered that glucose was mostly consumed after overnight culture and this is the reason that cells growth didn't observed and the absorbance values of DNS assay were almost zero.
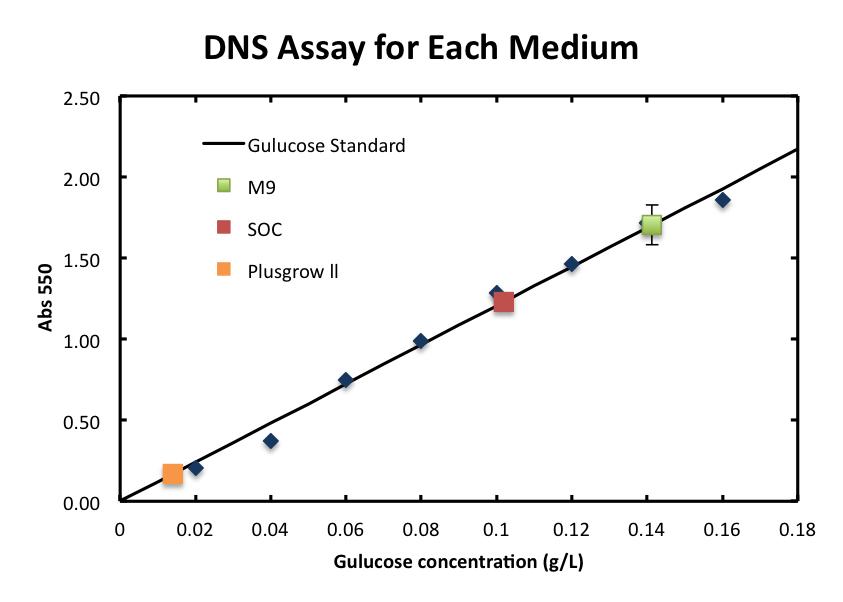
- 3-2-3 The DNS assay of medium.
- We checked the influence of each medium to the DNS assay. Figure 4 shows the background absorbance of each medium. The absorbance of M9 was 1.7±0.1, SOC was1.227±0.007, and Plusgrow Ⅱ was 0.17±0.02.
Discussion
M9 and SOC medium should be suit for the DNS assay using culture supernatant.
From three experiments: 2-1, 2, and 3, M9 and SOC are good media for characterizing the chitinase activity using the DNS assay. Firstly, cell number didn't increase so much about M9 and SOC. So, we can based on the premise that the enzyme concentration is nearly constant. Secondly, glucose or other materials in supernatant that react with DNS is almost consumed after overnight culture.
Using culture supernatant is available for the DNS assay.
From the data of Plusgrow Ⅱ on figure 2 and 3, the absorbance value of DNS assay didn't changed even if the cell number was increased; cells consumed sugar.
Reference
[1] “Actinobacteria.” Internet: http://en.wikipedia.org/wiki/Actinobacteria [Nov. 5, 2011]
[2] H. Ikeda, J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, S. Omura, “Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis.” Nat Biotechnol., vol. 21, no. 5 pp. 526-31, Apr. 2003
[3] S. Omura, H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, “Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites.” Proc Natl Acad Sci U S A. vol. 98, no. 21 pp. 12215-20, Oct. 9
[4] H. Sakuzou, ”還元糖の定量法(生物化学実験法)(kangento no teiryoho)” Kyoto University: Japan Scientific Soceties Press
[5] S. Kongruang, M. J. Han, C. I. Breton, M. H. Penner, “Quantitative Analysis of Cellulose-Reducing Ends.” Appl Biochem Biotechnol. Vol. 113, no. 116 pp. 213-31, Spring 2004
 "
"