Team:NYMU-Taipei/synthoprime
From 2011.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
---- | ---- | ||
<font size=3> | <font size=3> | ||
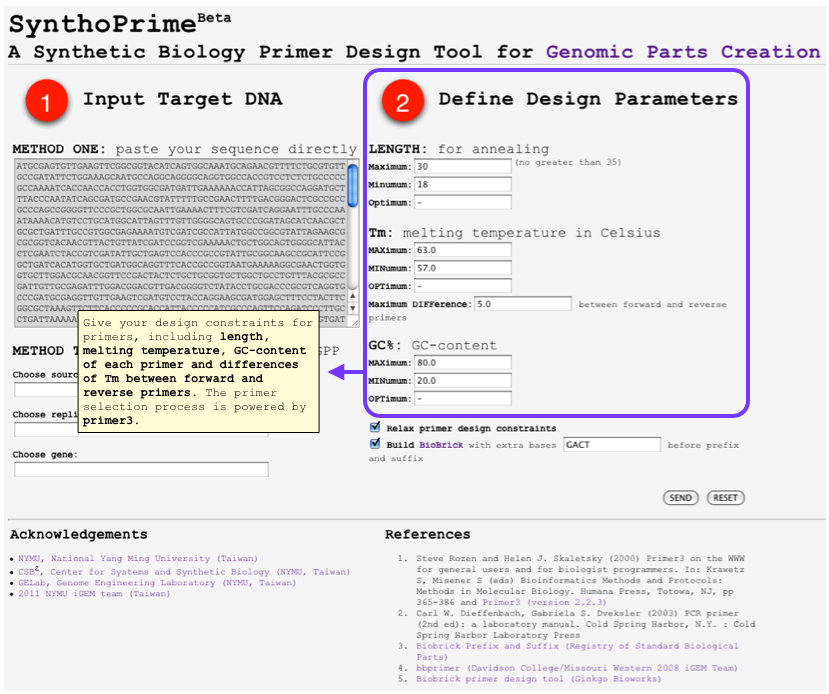
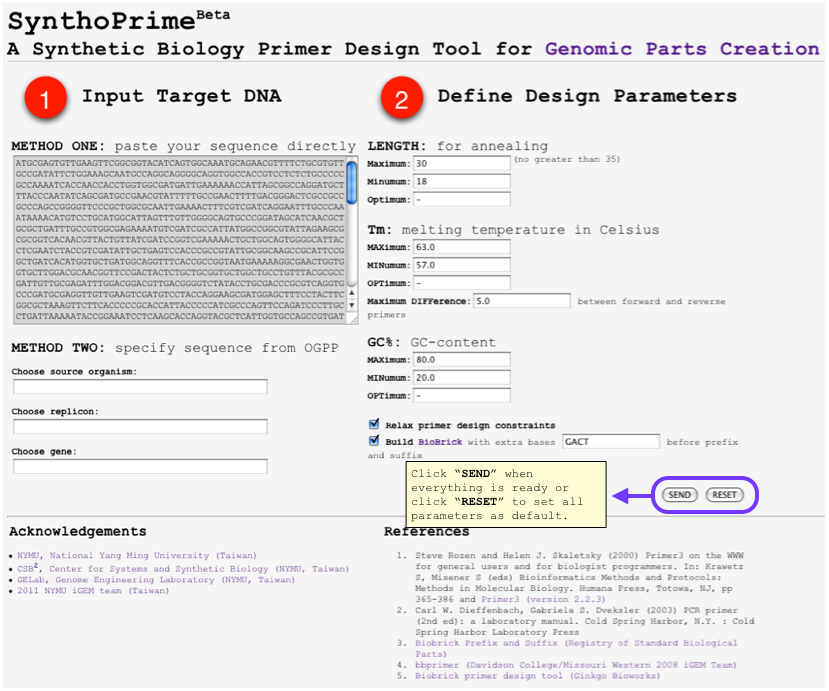
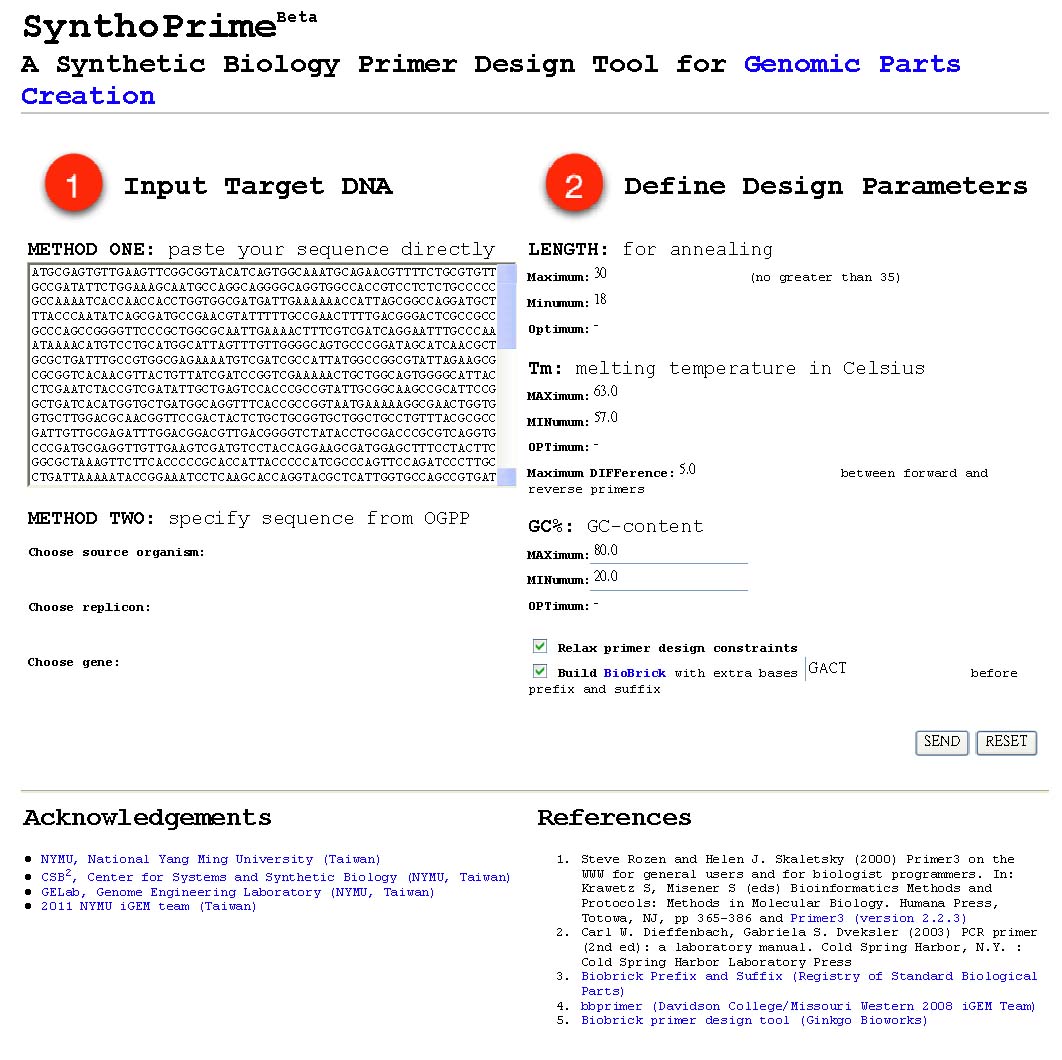
| - | *<font color=red>[http://120.126.44.58/SynthoPrime/ SynthoPrime]</font> algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. It provides an automated ONE-STEP primer design service of BioBricks for iGEM. Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community. [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out | + | *<font color=red>[http://120.126.44.58/SynthoPrime/ SynthoPrime]</font> algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. <font color=medium blue>It provides an automated ONE-STEP primer design service</font> of BioBricks for iGEM. <font color=medium blue>Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted</font> from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. <font color=medium blue>Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community.</font> [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
---- | ---- | ||
Revision as of 17:39, 5 October 2011


Contents |
Introduction
- [http://120.126.44.58/SynthoPrime/ SynthoPrime] – A Synthetic Biology Primer Design Tool for Genomic Parts Creation
 ]
]
- [http://120.126.44.58/SynthoPrime/ SynthoPrime] algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. It provides an automated ONE-STEP primer design service of BioBricks for iGEM. Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community. [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out.
HOWTO:User Inputs
HOWTO:Output Reading
References and Feedback
 "
"