Team:HKU-Hong Kong/Parts
From 2011.igem.org
| Line 15: | Line 15: | ||
|- | |- | ||
|style="width:900px;"| | |style="width:900px;"| | ||
| - | By expressing the tetR | + | By expressing the tetR-HNS fusion proteins, the tetR part of the proteins wound recognize and specifically bind to the 2 tetR binding region (tetO2).Thus in our designed operator- repressor binding, the presence of 2 tetR binding sites are used to enhance the binding of tetR part of the fusion proteins. As the fusion proteins bound to tetO, it is expected that the H-NS part of the fusion protein would attract and oligomerize with other native H-NS proteins inside the cell. With the oligomerization of the fusion proteins and native H-NS, the DNA covered by the oligomers is expected to trap the RNA polymerase during transcription, thus gene (GFP) repression is achieved. |
|- | |- | ||
|style="width:900px;"| | |style="width:900px;"| | ||
Revision as of 16:38, 5 October 2011
| Sample Data Page |
|
By expressing the tetR-HNS fusion proteins, the tetR part of the proteins wound recognize and specifically bind to the 2 tetR binding region (tetO2).Thus in our designed operator- repressor binding, the presence of 2 tetR binding sites are used to enhance the binding of tetR part of the fusion proteins. As the fusion proteins bound to tetO, it is expected that the H-NS part of the fusion protein would attract and oligomerize with other native H-NS proteins inside the cell. With the oligomerization of the fusion proteins and native H-NS, the DNA covered by the oligomers is expected to trap the RNA polymerase during transcription, thus gene (GFP) repression is achieved. |
|
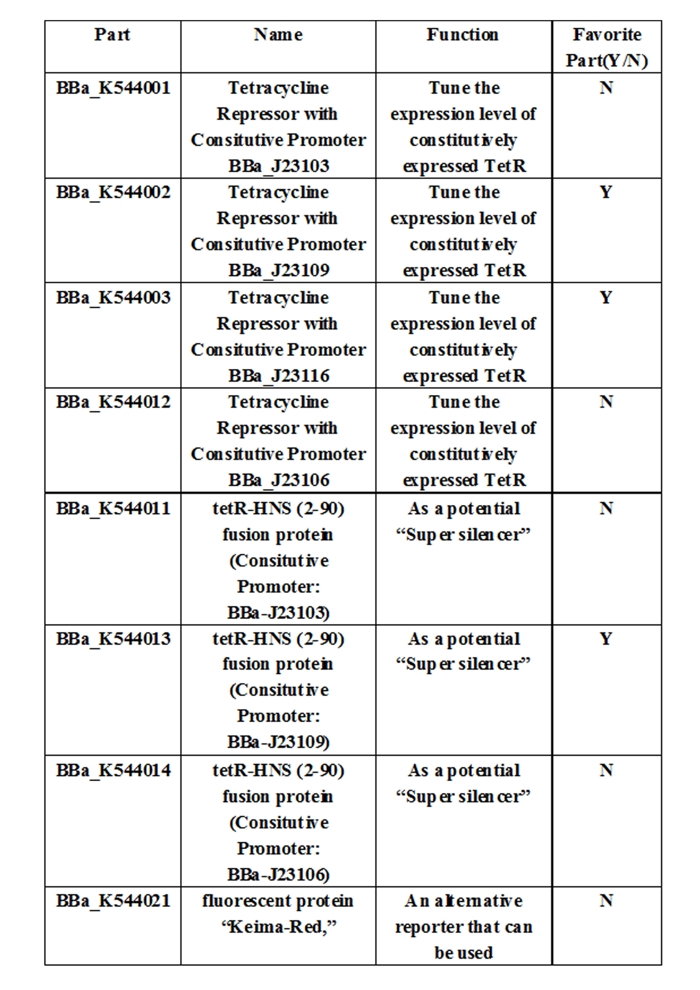
Experience: We have entered the confirmation data for the characterization of the selected constitutive promoters we have used. Of the 4 promoters we have used in the project, our team considered that J23109 or J23116 are more suitable promoters in creating the super silencer repression system. These parts are:
|
 "
"