Team:WHU-China/Standard
From 2011.igem.org
| Line 65: | Line 65: | ||
<div class="standard" id="standard2" > | <div class="standard" id="standard2" > | ||
| - | <p class="start">As we know | + | <p class="start">As we know, biobrick is a piece of DNA that can be spliced to any other biobrick. Standard biobricks share the same backbone with four restriction endonuclease sites: EcoRI & XbaI in the prefix and SpeI & PstI in the postfix. In traditional assembly, one biobrick is cut with E & S or X & P while the other is cut with E & X or S & P correspondingly. After mixing and ligation, the two biobricks are linked together, producing a new standard biobrick with the same backbone. There is always a resistance gene in each backbone as well, which gives the bacteria resistance against chloramphenicol, kanamycin, ampicillin or tetracycline resistance gene. The smurfly delicate design offers us great convenience. </br> |
| - | + | However, the standard assembly left a problem: there’s no way to break down the biobricks once they are assembled. We met that problem when we got a composite device including several biobricks from Edingburgh team. We tried to get a single biobrick which we needed from it and we had nothing to do but design a specific PCR reaction which cost time and money. </br> | |
| - | + | Therefore we hope to modify the standard backbone to make biobricks to better meet different requirements. | |
</p> | </p> | ||
<div class="pic" id="pic21"> | <div class="pic" id="pic21"> | ||
| Line 74: | Line 74: | ||
</div> | </div> | ||
<p class="con"> | <p class="con"> | ||
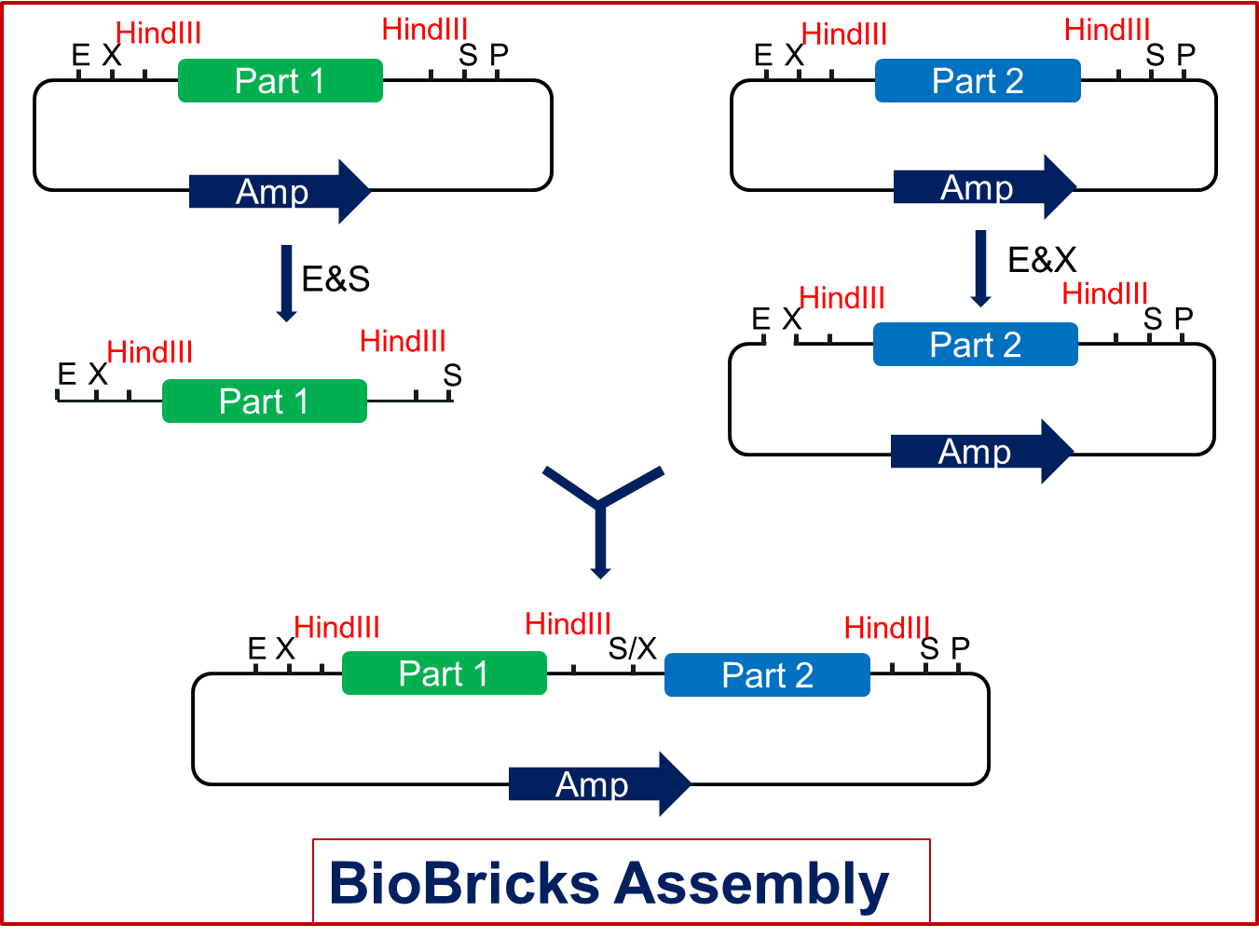
| - | Firstly, we | + | Firstly, we keep all the previous parts of the backbone so that the standard assembly still works for these new backbone plasmids (<span style="font-weight:bold">Fig2.1</span>). |
| - | Secondly, we | + | Secondly, we add another restriction endonuclease site--HindIII both in the prefix and postfix of the backbones (<span style="font-weight:bold">Fig2.1</span>). The order is shown in Fig2.1. Biobricks inserted in this kind of backbone can be assembled as the former standard. But once we produce the composite product, it will be different. Each part of the product will get two HindIII restriction sites, one upstream and the other downstream. |
</p> | </p> | ||
<div class="pic" id="pic22"> | <div class="pic" id="pic22"> | ||
| Line 82: | Line 82: | ||
</div> | </div> | ||
<p class="con"> | <p class="con"> | ||
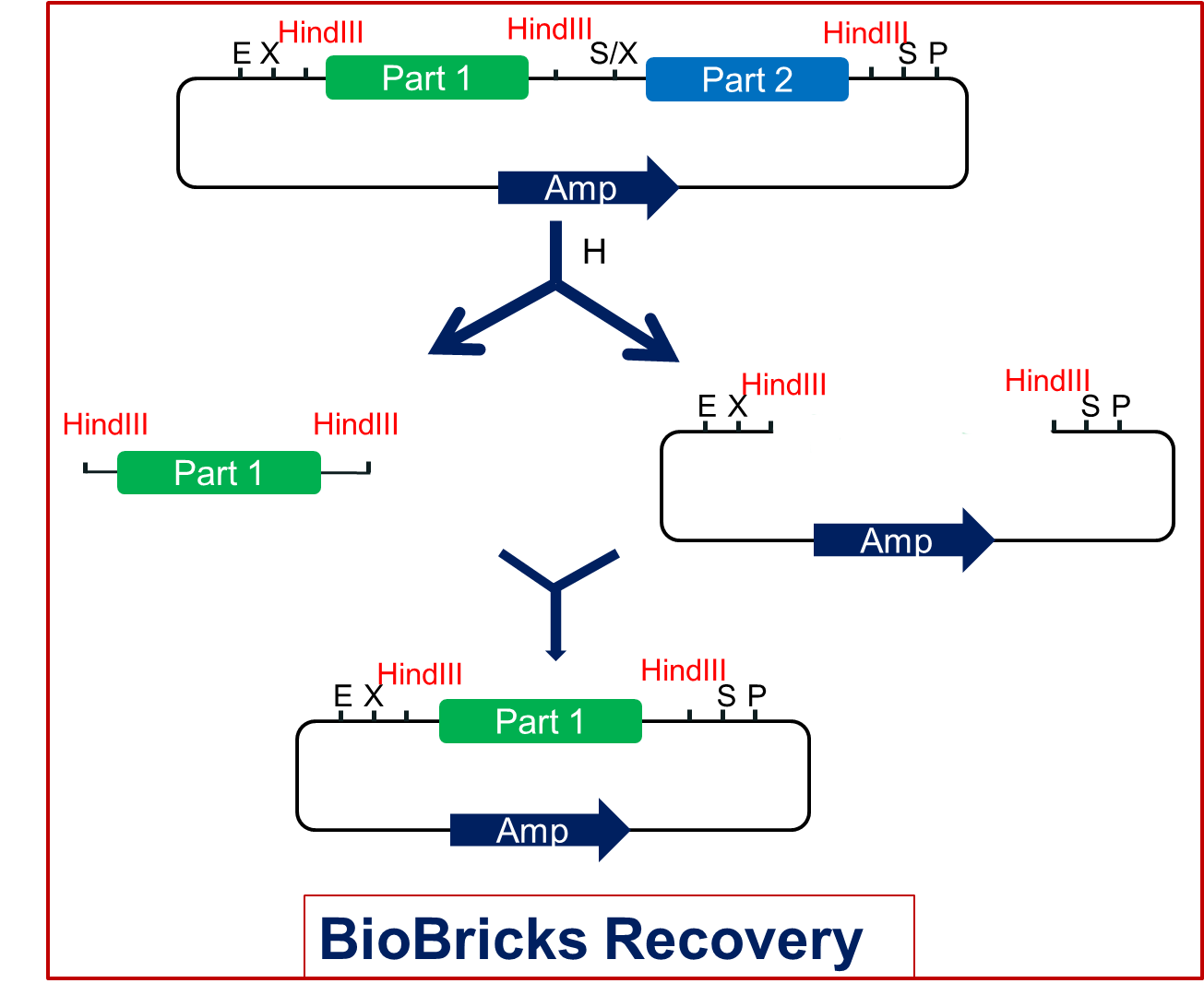
| - | + | In the new assembly, we can cut this product with the HindIII restriction endonuclease and get each part as well as the vector. Then we just need to link the part with the vector. Thus we can get the single standard biobrick of each part(<span style="font-weight:bold">Fig2.2</span>). | |
</p> | </p> | ||
<div class="pic" id="pic32"> | <div class="pic" id="pic32"> | ||
Revision as of 14:56, 3 October 2011
As we know, biobrick is a piece of DNA that can be spliced to any other biobrick. Standard biobricks share the same backbone with four restriction endonuclease sites: EcoRI & XbaI in the prefix and SpeI & PstI in the postfix. In traditional assembly, one biobrick is cut with E & S or X & P while the other is cut with E & X or S & P correspondingly. After mixing and ligation, the two biobricks are linked together, producing a new standard biobrick with the same backbone. There is always a resistance gene in each backbone as well, which gives the bacteria resistance against chloramphenicol, kanamycin, ampicillin or tetracycline resistance gene. The smurfly delicate design offers us great convenience. However, the standard assembly left a problem: there’s no way to break down the biobricks once they are assembled. We met that problem when we got a composite device including several biobricks from Edingburgh team. We tried to get a single biobrick which we needed from it and we had nothing to do but design a specific PCR reaction which cost time and money. Therefore we hope to modify the standard backbone to make biobricks to better meet different requirements.
Firstly, we keep all the previous parts of the backbone so that the standard assembly still works for these new backbone plasmids (Fig2.1). Secondly, we add another restriction endonuclease site--HindIII both in the prefix and postfix of the backbones (Fig2.1). The order is shown in Fig2.1. Biobricks inserted in this kind of backbone can be assembled as the former standard. But once we produce the composite product, it will be different. Each part of the product will get two HindIII restriction sites, one upstream and the other downstream.
In the new assembly, we can cut this product with the HindIII restriction endonuclease and get each part as well as the vector. Then we just need to link the part with the vector. Thus we can get the single standard biobrick of each part(Fig2.2).

 "
"

