Team:MIT/Notebook/
From 2011.igem.org
(Difference between revisions)
| Line 664: | Line 664: | ||
<h1>Week 5: July 4 - July 10</h1> | <h1>Week 5: July 4 - July 10</h1> | ||
We began looking into using the Goldengate assembly method, but two gene elements contained a cut site that needed to be mutated out. We performed Site-Directed Mutagenesis using the Lightning Kit in the hopes of mutating out the cut site, but results were not successful due to mishandling during the protocol, so the procedure was set to be repeated. Gibson assembly of AVPR2-TEVs-GV16 continues as we re-PCRed the AVPR2 DNA segment and re-run the entire Gibson protocol, picking 20 colonies in a determined attempt to obtain a successful result. | We began looking into using the Goldengate assembly method, but two gene elements contained a cut site that needed to be mutated out. We performed Site-Directed Mutagenesis using the Lightning Kit in the hopes of mutating out the cut site, but results were not successful due to mishandling during the protocol, so the procedure was set to be repeated. Gibson assembly of AVPR2-TEVs-GV16 continues as we re-PCRed the AVPR2 DNA segment and re-run the entire Gibson protocol, picking 20 colonies in a determined attempt to obtain a successful result. | ||
| + | <!-----------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | |||
| + | <> | ||
| + | </!> | ||
| + | |||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 670: | Line 677: | ||
<h1>Week 6: July 11 - July 17</h1> | <h1>Week 6: July 11 - July 17</h1> | ||
In the sixth week we continued re-attemping previous failures using modified protocols or higher quality DNA in hopes of obtaining successful reactions. We also began looking at Cadherins and the possibility of using them as a clumping mechanism for mammalian cells. In order to visualize Cadherins, we needed some sort of fluorescent color, so construction of NCadherin-EGFP (NCadherin is one of many types of cadherin) began. DNA for NCad-EGFP was ordered, but needed to be in a format such that we could LR react it with the different promoters we have. This is done by attaching attB sites to flank the NCad-EGFP gene and then performing the BP reaction, which generates LR reaction-compatible parts. PCR of the attB sites was successful. By this week we have also begun work with mammalian cell cultures. To explore the limitless possibilities of synthetic biology, a few of us took it upon themselves to look into other interesting gene components, such as the Caspase gene and the FF4 tag. | In the sixth week we continued re-attemping previous failures using modified protocols or higher quality DNA in hopes of obtaining successful reactions. We also began looking at Cadherins and the possibility of using them as a clumping mechanism for mammalian cells. In order to visualize Cadherins, we needed some sort of fluorescent color, so construction of NCadherin-EGFP (NCadherin is one of many types of cadherin) began. DNA for NCad-EGFP was ordered, but needed to be in a format such that we could LR react it with the different promoters we have. This is done by attaching attB sites to flank the NCad-EGFP gene and then performing the BP reaction, which generates LR reaction-compatible parts. PCR of the attB sites was successful. By this week we have also begun work with mammalian cell cultures. To explore the limitless possibilities of synthetic biology, a few of us took it upon themselves to look into other interesting gene components, such as the Caspase gene and the FF4 tag. | ||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | |||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 676: | Line 690: | ||
<h1>Week 7: July 18 - July 24</h1> | <h1>Week 7: July 18 - July 24</h1> | ||
Part of the team worked on creating protocols for a robot liquid handler to run the usual lab reactions that we run. We are hoping that the robot can replace us and do our liquid-related lab work for us. Various dry test runs were done. We transfected various DNA parts that we have into Hek293 cells, and results show that most of our DNA works. We investigated the TRE-rtTA3 system as well as the UAS-Gal4 system. Both seemed to be functional. | Part of the team worked on creating protocols for a robot liquid handler to run the usual lab reactions that we run. We are hoping that the robot can replace us and do our liquid-related lab work for us. Various dry test runs were done. We transfected various DNA parts that we have into Hek293 cells, and results show that most of our DNA works. We investigated the TRE-rtTA3 system as well as the UAS-Gal4 system. Both seemed to be functional. | ||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 682: | Line 701: | ||
<h1>Week 8: July 25 - July 31</h1> | <h1>Week 8: July 25 - July 31</h1> | ||
This week we underwent a momentous drive to create more LRs. A large list of promoter-gene pairs was conceived of and we began to run through LR reactions in somewhat of a factory manner. This occupied much of our time. We also received and prepared DNA parts from Elowitz's group in Caltech. We also got trained on using the FACS machine and began to get quantitative data on our transfections. | This week we underwent a momentous drive to create more LRs. A large list of promoter-gene pairs was conceived of and we began to run through LR reactions in somewhat of a factory manner. This occupied much of our time. We also received and prepared DNA parts from Elowitz's group in Caltech. We also got trained on using the FACS machine and began to get quantitative data on our transfections. | ||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | <!------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 688: | Line 713: | ||
<h1>Week 9: August 1 - August 7</h1> | <h1>Week 9: August 1 - August 7</h1> | ||
In the ninth week we did a lot of internal re-organization to increase our overall efficiency in work. This involved re-organizing an internal wiki that we use for management of our available DNA as well as a log of transfections needed to be done. Lots of samples were FACS-ed, generating lots of results that we can make graphs out of. Many of our parts were successfully characterized. | In the ninth week we did a lot of internal re-organization to increase our overall efficiency in work. This involved re-organizing an internal wiki that we use for management of our available DNA as well as a log of transfections needed to be done. Lots of samples were FACS-ed, generating lots of results that we can make graphs out of. Many of our parts were successfully characterized. | ||
| + | <!-------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | |||
| + | <!-------------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 696: | Line 728: | ||
<!-----------------------------------------------------------------------------------------------------------------> | <!-----------------------------------------------------------------------------------------------------------------> | ||
</!> | </!> | ||
| + | |||
| + | <h2>Week 10</h2> | ||
h3. August 14 | h3. August 14 | ||
| Line 754: | Line 788: | ||
Clara: Miniprepped and nanodropped 12 LRs from Aug 9. Hef1a: Delta-mCherry has esp. low concentration. To see conc. of other samples, refer to 8/11 [iGEM2011:Clara's Notebook] | Clara: Miniprepped and nanodropped 12 LRs from Aug 9. Hef1a: Delta-mCherry has esp. low concentration. To see conc. of other samples, refer to 8/11 [iGEM2011:Clara's Notebook] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 804: | Line 817: | ||
Charles: Completed Tyler's Hef1a_GV16 page with pretty FACS images. Watched Jenny do FACS at Koch. Some problems with either no cells in sample or visibly clumped cells. Introducing a new page...&nbsp;[iGEM2011:Color Palette] | Charles: Completed Tyler's Hef1a_GV16 page with pretty FACS images. Watched Jenny do FACS at Koch. Some problems with either no cells in sample or visibly clumped cells. Introducing a new page...&nbsp;[iGEM2011:Color Palette] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 921: | Line 909: | ||
<!----------------------------------------------------------------------------------------------------------------> | <!----------------------------------------------------------------------------------------------------------------> | ||
</!> | </!> | ||
| - | < | + | <h2> Week 11 </h2> |
h3. &nbsp;August 21 | h3. &nbsp;August 21 | ||
Revision as of 20:54, 28 September 2011
Overview
Here you can see what we did weekly in the lab!
Week 1: June 6 - June 11
In the first week we began constructing basic reporter DNA constructs that have various combinations of promoters and genes. The mix-and-matching of promoters and genes was done using the LR reaction following the Gateway protocol. Our promoters included TRE, minCMV, Hef1a, Hefl1a-LacO, and our genes included eYFP, mKate, and eBFP2. Transformed and inoculated colonies from the LR reactions and performed restriction digests. Approximately half of the LR reactions were successful and became DNA parts that we could use later on.June 7, 2011
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
C1434VP16 |
Yes. 6.7.2011, 5pm |
11 |
5, AMP, KH |
Standard, 30C for 16h |
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
Mnt1VP16 |
Yes. 6.7.2011, 5pm |
12 |
4, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Tyler |
2-3 |
Tre |
LacIKrab |
Yes. 6.7.2011, 5pm |
6 |
30, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
3-4 |
minCMV-7xMnt1 |
eYFP |
Yes. 6.7.2011, 5pm |
9 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
2-3 |
Tre |
LexAVP16 |
Yes. 6.7.2011, 5pm |
10 |
2, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 |
minCMV 1xC1434 |
eYFP |
Yes. 6.7.2011, 5pm |
7 |
3, AMP, KH, needs to be redone | Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 | minCMV 4xLexA |
eYFP |
Yes. 6.7.2011, 5pm |
8 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
LR Reactions
Utilizing protocol here: LR Protocol
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Grant |
3-4 |
minCMV 4xLexA | eYFP | Yes. 6.9.2011, 3pm |
12 |
10, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xMnt1 |
eYFP |
Yes. 6.9.2011, 3:45pm | 2 |
13, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xCI434 |
eYFP |
Yes. 6.9.2011, 3:45pm | 1 |
7, AMP, GR | Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
Hef1a-LacO |
eYFP |
Yes, 6.9.2011, 4:20pm |
5 |
25, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
4-5 |
Tre |
mKate |
Yes, 6.9.2011 4:27pm |
3 |
6, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
1-2 |
Hef1a |
eBFP2 |
Yes, 6.9.2011, 4.27pm |
4 |
3, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
Transformation
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Colony Number, Antibiotic |
Protocol Used | |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Mariola |
|
|
eYFP |
no. Replication stock. | |
2, AMP |
Transformation (~ 2 hr 20 min) using LB instead of SOC |
Miniprep
| Date | Assignee |
DNA |
Quantity |
Time collected |
|---|---|---|---|---|
| 6.9.2011 |
Divya-Jenny |
pEXPR_2-3_Tre:LexAVP16 |
52.2 ng/uL |
12pm |
| 6.9.2011 |
Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A) |
70 ng/ul |
12pm |
| 6.9.2011 |
Kenneth | pEXPR_3-4_minCMV-CI434:eYFP (B) |
174 ng/uL |
12pm |
| 6.9.2011 |
Louis |
pEXPR_2-3_Tre:C1434VP16 |
234.6 ng/uL |
12pm |
| 6.9.2011 |
Tyler |
pEXPR_2-3_Tre:Lac/Krab |
130.7 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
110.0 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pDEST_2-3_ccdB |
117.6 ng/uL |
12pm |
Restriction Digests
| Assignee |
DNA |
Enzyme |
Expected Results |
Picture of Gel |
Time Incubated | Comments |
||
|---|---|---|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | NdeI |
6700 bp 800 bp |
a.3 | 6/9/11 3:50 PM |
|
||
| Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A+B) |
NcoI and SacII |
2450 bp 4650 bp |
A: a.6 B: a.7 |
6/9/11 3:00 PM |
A did not cut as expected. B looks good. Digestion Mix: 2 uL NEB4, 0.5 uL of each enzyme, A: 7.1 uL/B: 2.9 uL of DNA, 10.4 uL/14.6 uL of H20 |
||
| Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
BglI |
3700 bp 2200 bp 1300 bp |
a.1 | 6/9/11 4:00PM |
DNA appeared to be uncut. Need to redo, possibly with a new enzyme and more DNA. |
||
| Michelle |
pDEST_2-3_ccdB |
NcoI and NheI |
6100 bp 1700 bp |
a.2 | 6/9/2011 4:25 PM |
Attained bands at correct positions. Other band represents partially cut DNA in double digest. |
||
| Divya |
pEXPR_2-3_Tre:LexAVP16 |
HincII |
a.4 | 6/9/2011 4:45 PM |
- Bands didn't show up. Needs to be redone. - DNA may have degraded. Need to redo Nanodrop as well. |
|||
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
a.5 | 6/9/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL Only saw one band at 3500 bp Needs to be redone possibly with another restriction enzyme |
Gels
June 10, 2011
Restriction Gels
| Assignee | DNA |
Enzyme |
Expected result |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | EcoRI (Buffer 2) |
5500 bp 1050 bp 650 bp 360 bp |
6/10/11 11:15 AM |
|
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (A) |
NcoI and NheI |
5700 bp 1300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NcoI 1 uL Nhe1 2 uL NEB2 2 uL BSA 8 uL H2O Failed. |
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (B) |
NheI and SphI |
1700 bp 5300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NheI 1 uL SphI 2 uL NEB2 2 uL BSA 8 uL H2O Failed. Redoing LR on Monday June 13th. |
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
6/10/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL ***Worked (see gel below - b.1) Stored in -80C freezer |
Comments:
pEXPR_2-3_Tre:LexAVP16 and pEXPR_3-4_minCMV-7xMnt1:eYFP are being entirely redone.
pEXPR_2-3_Tre:LexAVP16 (post-miniprep) was lost. pEXPR_3-4_minCMV-7xMnt1:eYFP LR was unsuccessful (no bacteria grew).
LR and transformation will be done on Saturday. Miniprep and restriction mapping will be done on Sunday
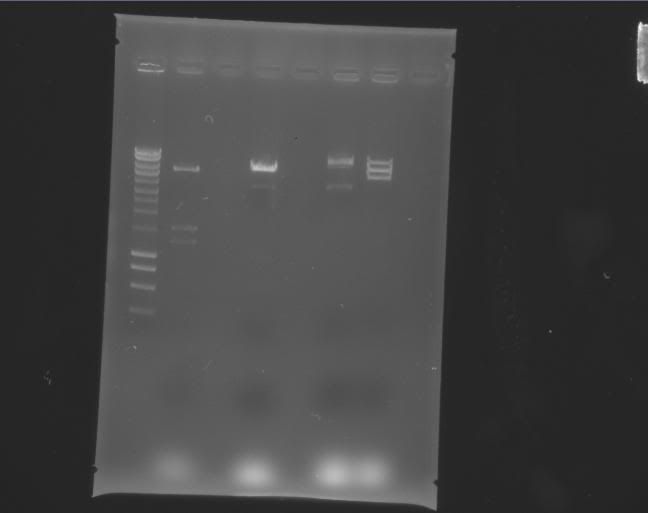
| Label | Gel |
Legend |
|---|---|---|
| b |
 > > |
Column 0: HyperLadder Column 1: pEXPR_2-3_Tre:Lac/Krab Column 4: pEXPR_2-3_Tre:C1434VP16 Column 6: pEXPR_2-3_TRE:Mnt1VP16 (A) Column 7: pEXPR_2-3_TRE:Mnt1VP16 (B) |
June 11, 2011
Set of LRs from 6/9 (GR, JC, and SR) cell counts taken; inoculated by GR/JC. 3 cells taken per plate.
| Assignee |
DNA |
Enzyme |
Expected Results |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Sam |
pEXPR_1-2_Hef:eBFP |
NcoI & ApaL1 |
3750bp, 3180bp, 1250bp | 45 min | If it doesn't work there should be a whole mess of fragments (including a small 600bp one) 1mL each enz; 2 mL Buf; 1 mL BSA; 2mL DNA; 13mL H2O (Standard Protocol with extra BSA) |
| Sam |
pEXPR_4-5_Tre:mKate |
Bgl1 |
1270bp, 2630bp, 3380bp | 45 min | 1mL enz; 2mL DNA; 2mL BSA; 2mL Buf; 15mL H2O (Standard Protocol with extra BSA and water - not quite 10x) |
 "
"