Team:MIT/Notebook/
From 2011.igem.org
(Difference between revisions)
(→Overview) |
|||
| Line 694: | Line 694: | ||
<h1>Week 10: August 8 - August 14</h1> | <h1>Week 10: August 8 - August 14</h1> | ||
Computer simulations of the Notch-Delta interactions were presented in our group this week, and we became convinced of the possibility of creating self-patterning mammalian cells. On the DNA side of things, we are trying to create more and more DNA using miniprep, because midipreps have for some reason not been successful for us or the 2010 iGEM team. On the transfection side, a lot of new DNA parts were transfected, observed under the microscope, and FACS-ed to quantify their effect/behavior. | Computer simulations of the Notch-Delta interactions were presented in our group this week, and we became convinced of the possibility of creating self-patterning mammalian cells. On the DNA side of things, we are trying to create more and more DNA using miniprep, because midipreps have for some reason not been successful for us or the 2010 iGEM team. On the transfection side, a lot of new DNA parts were transfected, observed under the microscope, and FACS-ed to quantify their effect/behavior. | ||
| + | <!-----------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | h3. August 14 | ||
| + | |||
| + | |||
| + | |||
| + | Charles - Miniprepped propagations of various things, good CP LRs, as well as re-inoculations of poor CP LRs. Nanodrops were all over hte place. Some of the re-inoculated CP LRs reached 500 ng/uL, which was weird. &nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Divya - Ran FACS on 50 samples for Tyler and Ken. Also, I now have Koch access\! Woohoo\!\!\! | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth - Prepped FACS samples. Moved frozen cells to ln2 | ||
| + | |||
| + | |||
| + | |||
| + | Grant - Today and yesterday did sequencing of Tango and other plasmids as well as propagations of a handful of failed plasmids / spinning down of cells for Charles' miniprep. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 13 | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth - Added delta to cells. Co-cultured AVPR and pDisplay Vasopressin cells. Froze down and passed the Elowitz CHO cell lines and put in \-80. Also passed wells and flasks. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 12 | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth - Transfected cells for AVPR2 experiment as well as another free delta induction experiment, this time with const. color gating. | ||
| + | |||
| + | |||
| + | |||
| + | Charles - Attended Ken's orientation session and did 1 transfection just to try things out. Also obtained my new culture of cells. Cell stocked CP LR 4,5,6,7,8,11,12, of which 4,5, and 7 are tentative (repeat submitted). Re-inoculated failed CP LR's 1,2,3,9,10 with a different colony. Also propagated a whole bunch of plasmids, including the successful and tentatively successful CP LR's. Hopefully the color palette will be complete by Tuesday. &nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Jon - Tissue culture orientation & inoculated SDM transformations from yesterday. Colony counts around 20 for TEV and 3 for GV16. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | h3. August 11 | ||
| + | |||
| + | |||
| + | |||
| + | Jon - Redid GV16 and TEV SDM with new primers; transformed, hopefully they work this time. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Clara: Miniprepped and nanodropped 12 LRs from Aug 9. Hef1a: Delta-mCherry has esp. low concentration. To see conc. of other samples, refer to 8/11 [iGEM2011:Clara's Notebook] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Tyler: Prepared cells for transfection tomorrow. Plan on doing the following experiments: NCAD phenotype, TRE:LacI and TRE:LacI-Krab with proper gating, Hef1A:GV16 with proper gating, DRD2 repeat, and possible CCL5 experiment. Did FACS on&nbsp;[iGEM2011:Confluency Experiment]&nbsp;which shows best starting value around 1.5*10^5 cells/mL starting concentration. FACS machine showed around 40% efficiency. Matlab shows around 30%. In either case, we should start transfections with cells less confluent than we have been. TRE:LacI-Krab experiment seems to have worked, but there is only about 2x repression (see bottom of&nbsp;[Tri-Color Experiment v2|iGEM2011:Tri-Color Experiment v2]). Also put up FACS from AmCyan which is more green than blue. | ||
| + | |||
| + | |||
| + | |||
| + | Michelle: Prepped Clara's 12 minipreps for sequencing. Worked on Circuit Diagrams and write ups for the public wiki. | ||
| + | |||
| + | |||
| + | |||
| + | Charles: 42 FACS in less than 1 hour. Very fast because Ken's samples were concentrated. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | h3. August 10 | ||
| + | |||
| + | |||
| + | |||
| + | Tyler: I updated the wiki (first (per usual(winning))). Prepared for FACS. Transfected Confluency Experiment ([iGEM2011:Confluency Experiment]). Will FACS tomorrow. Still need FACS data from Divya for Tre:LacI-Krab experiment and some AmCyan colors. Today Jenny FACSed some DRD2 (apparently a failure from pictures) and some Delta-Notch stuff (also a failure). Need to look at the FACS, which apparently disappeared today? so hopefully we can find that tomorrow as well. Will try DRD2 (+100 uM dopamine) again later in the week with possibly more dopamine.&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Charles: Completed Tyler's Hef1a_GV16 page with pretty FACS images. Watched Jenny do FACS at Koch. Some problems with either no cells in sample or visibly clumped cells. Introducing a new page...&nbsp;[iGEM2011:Color Palette] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Michelle: Inoculated Clara's LRs. Ready to miniprep and send for sequencing tomorrow. | ||
| + | |||
| + | |||
| + | |||
| + | Grant: Miniprepped, restriction mapped, and sequenced the minCMV promoters and a few other genes/promoters. Started LR reactions for a few more Tango parts and the Ephrin parts. Shockingly did nothing else. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 9 | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth: Worked on MATLAB stuff more. Treated TRE:mKate and TRE:eBFP2 experiments with dox. Transfected for pDisplay immunofluoresence experiment again. Thawed out Elowitz cell lines. Froze down large numbers of HEK and CHO cells. Passed cells into 60 mm plates for N-cad knockdown experiment. FACS data analysis. Prepared dopamine aliquots and supplies for future experiments. | ||
| + | |||
| + | |||
| + | |||
| + | Grant: Removed propagations of various gene/promoter entry vectors and inoculated a single colony from each. Did alignments of sequencing data for samples sent out yesterday. One LR reaction failed unexpectedly, but the other failures involving minCMV promoters appear to have occurred due to contamination from a Hef1a variant promoter based on a BLAST of the sequencing results. (READ: The "minCMV" promoter DNA is not actually minCMV promoter DNA\- at least this is the case for the tubes without stickers.) All parts currently being used for experiments are verified. On track for \~15-20 new LR reactions for Wednesday afternoon. | ||
| + | |||
| + | |||
| + | |||
| + | Clara: 12 LR reactions, left at room temperature @ 12:30pm~. Refer to [iGEM2011:Clara's Notebook]&nbsp;for details. | ||
| + | |||
| + | |||
| + | |||
| + | Charles: Transformed Clara's LR reactions. Had to leave during the middle of the day to go to the Student Services Office. Watched Mariola's presentation. Working on a new page. | ||
| + | |||
| + | |||
| + | |||
| + | Jenny: TriColor FACS are now up on their appropriate pages. Rest are temporarily on my notebook. | ||
| + | |||
| + | |||
| + | |||
| + | Tyler - Updated the wiki. Need to redo all of the FACS data tomorrow that Jenny did today because the red and blue colors are switched. Mariola's program works so I will use that. Tomorrow I will do transfections of a consistuitive color into wells with different confluencies that I set up today. I added Dox and Dopamine to wells today that we will FACS tomorrow.&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Michelle/Divya: Miniprepped and nanodropped 12 samples. Prepped other samples for sequencing. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | h3. August 8 | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth: planned out and did transfections for TRE eBFP2, TRE:mKate and AVPR2 validation experiments. Also threw in a co-culture of Notch/Delta cells, only with UAS:eYFP instead of UAS:citrine from Elowitz. Not expecting anything from that, but just covering all bases. Treated pDisplay-vasopressin-myc cells with anti-myc FITC Ab and prepared slide. Results suggestive. Cells were too confluent at time of fixation, many were washed off and the remaining look bad. Probably from my bootleg formaldehyde solution as well. However, remaining cells were green on the surface as well as red from delta-mcherry, while controls are not green. Seems to suggest it is being displayed... | ||
| + | |||
| + | |||
| + | |||
| + | Grant: Miniprepped the EPHB2, EFNB1, and CXCR1-TEVs-GV16 pENTR vectors assembled previously. Set up fourteen sequencing reactions of multiple Tango plasmids and a few reporter expression vectors designed to test the minCMV promoters, as well as the three ENTR vectors miniprepped. Pending affirmative sequencing results, LR reactions will begin. Because we need a number of promoters propagated and it would be nice to do LRs with the Rheoswitch system (and possibly the new Notch constructs) concurrently, I will probably wait to begin these. Cleaned the lab before the start of the work day, like a boss, or perhaps a janitor. Started propagations of a few promoters and genes that we had < 1 uL of DNA left of (and no cell stock), because if you want something done... | ||
| + | |||
| + | |||
| + | |||
| + | Tyler: Transfections | ||
| + | |||
| + | |||
| + | |||
| + | Charles: Propagated DNA we are low on. Looks like Grant also did this. So we should be stocked. And miscellanea. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Mariola: Aliquoted and Transfected UAS:EGFP and TRE:eyfp-4xFF4 experiments. Dox ladder of each. Writing 8pg REU paper and presentation. | ||
| + | |||
| + | |||
| + | |||
| + | DMC: Prepped and submitted several samples for sequencing. See Divya's Personal Notebook. Brought presents from Weiss Lab (SalI-HF, 10uL tips, 1000uL tips).&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {cloak} | ||
| + | |||
| + | <!-----------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
| Line 700: | Line 919: | ||
<h1>Week 11: August 15 - August 21</h1> | <h1>Week 11: August 15 - August 21</h1> | ||
In our experimental attempt to characterize the Notch-Delta interaction, we used co-culture of CHO and Hek293 cells as well as stable cell lines of Notch-containing and Delta-containing cells from Elowitz to observe the trans-activation of Notch by Delta. Other experiments using CHO cells, which are more difficult to transfect than Hek293 using Lipofectamine, were carried out to observe the behavior of CHO transfected cells. | In our experimental attempt to characterize the Notch-Delta interaction, we used co-culture of CHO and Hek293 cells as well as stable cell lines of Notch-containing and Delta-containing cells from Elowitz to observe the trans-activation of Notch by Delta. Other experiments using CHO cells, which are more difficult to transfect than Hek293 using Lipofectamine, were carried out to observe the behavior of CHO transfected cells. | ||
| + | <!----------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
| + | |||
| + | <!----------------------------------------------------------------------------------------------------------------> | ||
| + | </!> | ||
| + | |||
<div class="clear"></div> | <div class="clear"></div> | ||
</div> | </div> | ||
Revision as of 20:40, 28 September 2011
Overview
Here you can see what we did weekly in the lab!
Week 1: June 6 - June 11
In the first week we began constructing basic reporter DNA constructs that have various combinations of promoters and genes. The mix-and-matching of promoters and genes was done using the LR reaction following the Gateway protocol. Our promoters included TRE, minCMV, Hef1a, Hefl1a-LacO, and our genes included eYFP, mKate, and eBFP2. Transformed and inoculated colonies from the LR reactions and performed restriction digests. Approximately half of the LR reactions were successful and became DNA parts that we could use later on.June 7, 2011
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
C1434VP16 |
Yes. 6.7.2011, 5pm |
11 |
5, AMP, KH |
Standard, 30C for 16h |
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
Mnt1VP16 |
Yes. 6.7.2011, 5pm |
12 |
4, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Tyler |
2-3 |
Tre |
LacIKrab |
Yes. 6.7.2011, 5pm |
6 |
30, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
3-4 |
minCMV-7xMnt1 |
eYFP |
Yes. 6.7.2011, 5pm |
9 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
2-3 |
Tre |
LexAVP16 |
Yes. 6.7.2011, 5pm |
10 |
2, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 |
minCMV 1xC1434 |
eYFP |
Yes. 6.7.2011, 5pm |
7 |
3, AMP, KH, needs to be redone | Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 | minCMV 4xLexA |
eYFP |
Yes. 6.7.2011, 5pm |
8 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
LR Reactions
Utilizing protocol here: LR Protocol
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Grant |
3-4 |
minCMV 4xLexA | eYFP | Yes. 6.9.2011, 3pm |
12 |
10, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xMnt1 |
eYFP |
Yes. 6.9.2011, 3:45pm | 2 |
13, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xCI434 |
eYFP |
Yes. 6.9.2011, 3:45pm | 1 |
7, AMP, GR | Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
Hef1a-LacO |
eYFP |
Yes, 6.9.2011, 4:20pm |
5 |
25, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
4-5 |
Tre |
mKate |
Yes, 6.9.2011 4:27pm |
3 |
6, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
1-2 |
Hef1a |
eBFP2 |
Yes, 6.9.2011, 4.27pm |
4 |
3, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
Transformation
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Colony Number, Antibiotic |
Protocol Used | |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Mariola |
|
|
eYFP |
no. Replication stock. | |
2, AMP |
Transformation (~ 2 hr 20 min) using LB instead of SOC |
Miniprep
| Date | Assignee |
DNA |
Quantity |
Time collected |
|---|---|---|---|---|
| 6.9.2011 |
Divya-Jenny |
pEXPR_2-3_Tre:LexAVP16 |
52.2 ng/uL |
12pm |
| 6.9.2011 |
Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A) |
70 ng/ul |
12pm |
| 6.9.2011 |
Kenneth | pEXPR_3-4_minCMV-CI434:eYFP (B) |
174 ng/uL |
12pm |
| 6.9.2011 |
Louis |
pEXPR_2-3_Tre:C1434VP16 |
234.6 ng/uL |
12pm |
| 6.9.2011 |
Tyler |
pEXPR_2-3_Tre:Lac/Krab |
130.7 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
110.0 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pDEST_2-3_ccdB |
117.6 ng/uL |
12pm |
Restriction Digests
| Assignee |
DNA |
Enzyme |
Expected Results |
Picture of Gel |
Time Incubated | Comments |
||
|---|---|---|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | NdeI |
6700 bp 800 bp |
a.3 | 6/9/11 3:50 PM |
|
||
| Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A+B) |
NcoI and SacII |
2450 bp 4650 bp |
A: a.6 B: a.7 |
6/9/11 3:00 PM |
A did not cut as expected. B looks good. Digestion Mix: 2 uL NEB4, 0.5 uL of each enzyme, A: 7.1 uL/B: 2.9 uL of DNA, 10.4 uL/14.6 uL of H20 |
||
| Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
BglI |
3700 bp 2200 bp 1300 bp |
a.1 | 6/9/11 4:00PM |
DNA appeared to be uncut. Need to redo, possibly with a new enzyme and more DNA. |
||
| Michelle |
pDEST_2-3_ccdB |
NcoI and NheI |
6100 bp 1700 bp |
a.2 | 6/9/2011 4:25 PM |
Attained bands at correct positions. Other band represents partially cut DNA in double digest. |
||
| Divya |
pEXPR_2-3_Tre:LexAVP16 |
HincII |
a.4 | 6/9/2011 4:45 PM |
- Bands didn't show up. Needs to be redone. - DNA may have degraded. Need to redo Nanodrop as well. |
|||
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
a.5 | 6/9/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL Only saw one band at 3500 bp Needs to be redone possibly with another restriction enzyme |
Gels
June 10, 2011
Restriction Gels
| Assignee | DNA |
Enzyme |
Expected result |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | EcoRI (Buffer 2) |
5500 bp 1050 bp 650 bp 360 bp |
6/10/11 11:15 AM |
|
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (A) |
NcoI and NheI |
5700 bp 1300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NcoI 1 uL Nhe1 2 uL NEB2 2 uL BSA 8 uL H2O Failed. |
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (B) |
NheI and SphI |
1700 bp 5300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NheI 1 uL SphI 2 uL NEB2 2 uL BSA 8 uL H2O Failed. Redoing LR on Monday June 13th. |
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
6/10/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL ***Worked (see gel below - b.1) Stored in -80C freezer |
Comments:
pEXPR_2-3_Tre:LexAVP16 and pEXPR_3-4_minCMV-7xMnt1:eYFP are being entirely redone.
pEXPR_2-3_Tre:LexAVP16 (post-miniprep) was lost. pEXPR_3-4_minCMV-7xMnt1:eYFP LR was unsuccessful (no bacteria grew).
LR and transformation will be done on Saturday. Miniprep and restriction mapping will be done on Sunday
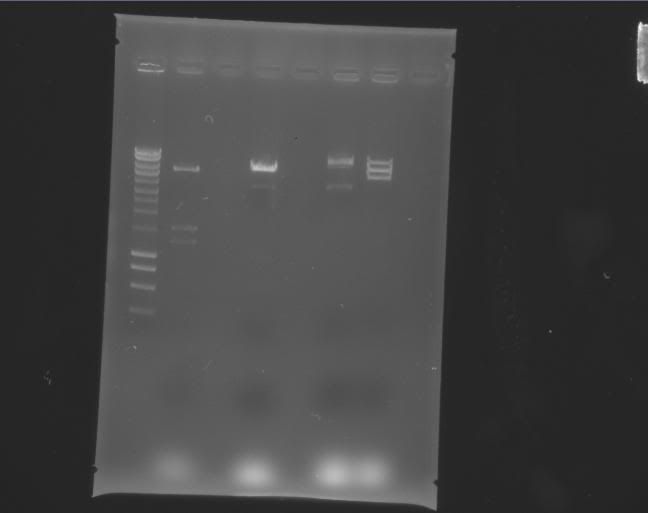
| Label | Gel |
Legend |
|---|---|---|
| b |
 > > |
Column 0: HyperLadder Column 1: pEXPR_2-3_Tre:Lac/Krab Column 4: pEXPR_2-3_Tre:C1434VP16 Column 6: pEXPR_2-3_TRE:Mnt1VP16 (A) Column 7: pEXPR_2-3_TRE:Mnt1VP16 (B) |
June 11, 2011
Set of LRs from 6/9 (GR, JC, and SR) cell counts taken; inoculated by GR/JC. 3 cells taken per plate.
| Assignee |
DNA |
Enzyme |
Expected Results |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Sam |
pEXPR_1-2_Hef:eBFP |
NcoI & ApaL1 |
3750bp, 3180bp, 1250bp | 45 min | If it doesn't work there should be a whole mess of fragments (including a small 600bp one) 1mL each enz; 2 mL Buf; 1 mL BSA; 2mL DNA; 13mL H2O (Standard Protocol with extra BSA) |
| Sam |
pEXPR_4-5_Tre:mKate |
Bgl1 |
1270bp, 2630bp, 3380bp | 45 min | 1mL enz; 2mL DNA; 2mL BSA; 2mL Buf; 15mL H2O (Standard Protocol with extra BSA and water - not quite 10x) |
 "
"