Team:NYC Software/Notebook/Summary
From 2011.igem.org
(Difference between revisions)
RussellDurrett (Talk | contribs)
(Created page with "{{:Team:NYC_Software/Templates/nav}} {{:Team:NYC_Software/Templates/css}} <html> <html lang="en"> <head> <meta charset="utf-8"> <title>NYC-iGEM software</title> <meta name "desc...")
Newer edit →
(Created page with "{{:Team:NYC_Software/Templates/nav}} {{:Team:NYC_Software/Templates/css}} <html> <html lang="en"> <head> <meta charset="utf-8"> <title>NYC-iGEM software</title> <meta name "desc...")
Newer edit →
Revision as of 18:57, 28 September 2011

Week--0
--Our first meeting of the summer is scheduled for TUESDAY, May 31st at NOON in the ICB Conference room here at Weill Cornell--LOL, Russell isn't an old dude
Week--1
--Familiarizing myself with the BioInformatics Toolbox in MATLAB - Balvir--Starting out learning Python, reviewing Russ's Python book & old scripts - Hannah
--This week code, next week genomics! - Russ
Week--2
-06/13/2011 - 06/17/2011--Many, many research papers on Whole Genome Analysis techniques to read
--Many, many software packages and tools needed to be downloaded onto my computer
Week--3
-06/20/2011 - 06/24/2011--Reproduced Russell's files for myself using PHYLIP and MATLAB - Balvir
--Finally got my ID. Still no Cluster access though.
Week--4
-06/27/2011 - 07/01/2011--Trying to figure out how to use the cluster. Whoooo!
--WTF this Unix book is thick and the font's too small
--But now you can be learning code on the subway! -Russ
Week--5
-07/05/2011 - 07/08/2011--Determined that FFP scripts should utilize an l-mer length of 19 based on relative entropy and max vocab values
--Generated a phylogenetic tree of the six selected species so far using the FFP outputs -Balvir -> now to visualize
--Conference call on Friday 07/08/2011
Week--6
-07/11/2011 - 07/15/2011--I imported Balvir's Trees into Geneious and formatted them to look pretty. Colored them so it's clear that they all differ in clades. Concatenated them all into one image and added to Previous Studies page but also here: -Russ
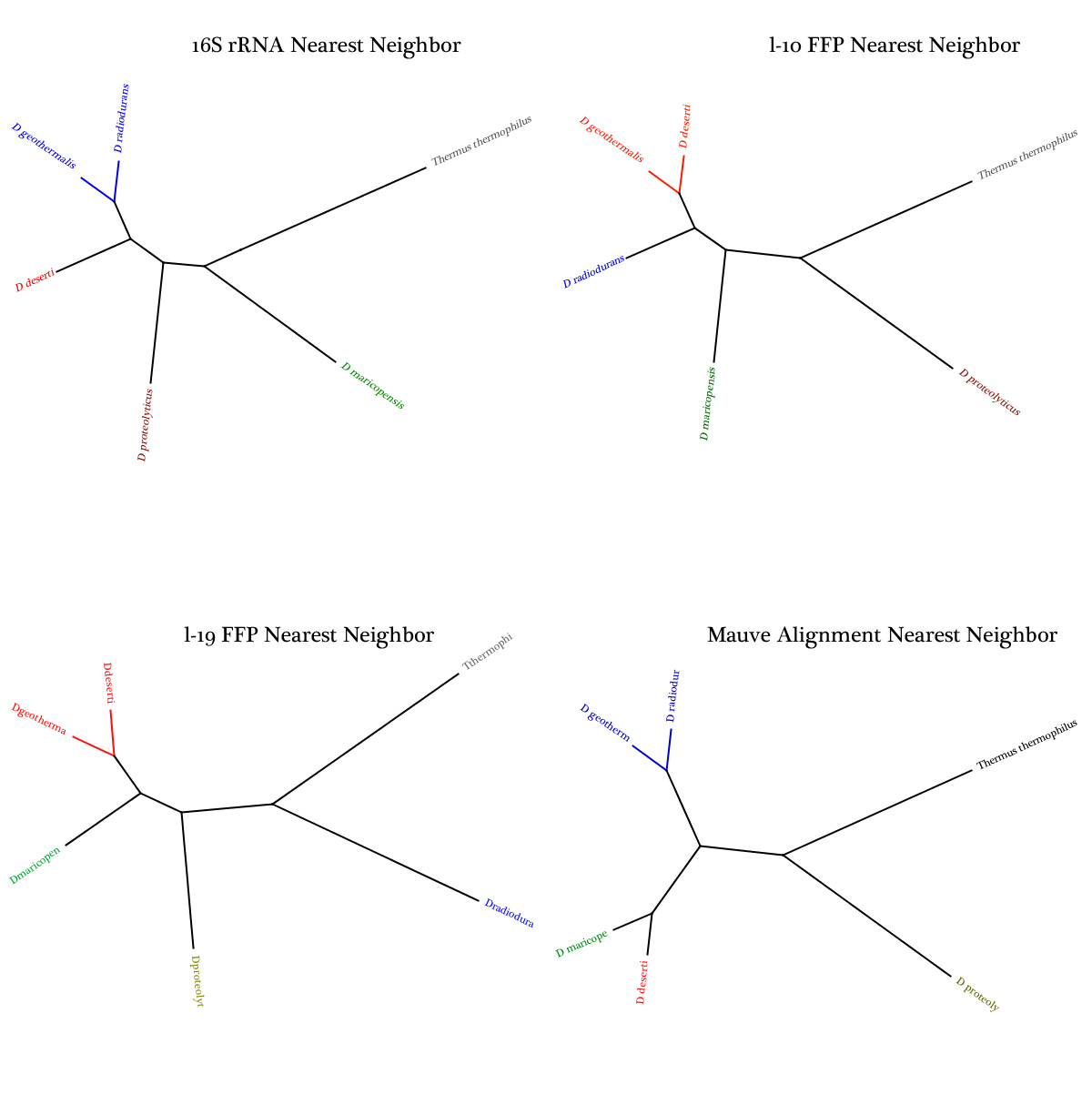
Week--7
-07/18/2011 - 07/22/2011--Now that you know some unix, let's explore genomic bioinformatics tools! Everybody download the BWA aligner and glimmer (or just RSYNC my bin on the cluster) -Russ
--Genomics is hard.
Week--8
-07/25/2011 - 07/29/2011--Downloaded the existing Deinococcus genomes, starting to use glimmer to get genes.
--Minimal Primer Designer is running - still need to add a lot of bells a whistles. - Hannah
Week--9
--08/01/2011 - 08/05/2011--One month to go, it's going to be tight. We received the pellets from Dr. Daly's group so I will begin extracting DNA and making sequencing libraries.
--Primer Designer working predictably on the command line. Tested the outputs in Geneious and it looks like it'll work. btw Geneious could use some updating as far as cloning primers go - Hannah
Week--10
--08/08/2011 - 08/12/2011--5 out of 8 DNA preps worked well enough to continue - I think there are a lot of carbs in these strains. I'll redo the others but move forward making libraries with these five. -Russ
--MatLab GUI-based BLAST script will be hard to port to the nysynbio.org server - learning python and redoing it. Ugh. -Balvir
Week--11
--08/15/2011 - 08/19/2011--Interesting news - poster was accepted at a conference in switzerland next week, so I'll be gone for 10 days. 5 libraries are prepared and will be submitted to the core for sequencing. We'll figure out how to get data from them later. -Russ
--Python BLAST Script is working now. -Balvir
Week--12
--08/22/2011 - 08/26/2011--Scripts are working now we just need to get them online. NYSynbio.org is being funky with scripts, so we may have to use russelldurrett.org. Sung volunteers to get them running while I'm away. See you all in two weeks! -Russ
 "
"