Team:Calgary/Project/Chassis
From 2011.igem.org
Emily Hicks (Talk | contribs) |
Emily Hicks (Talk | contribs) (P) |
||
| Line 22: | Line 22: | ||
</html>[[Image:UofC2011_OriT_oriV.png|frame|center|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> | </html>[[Image:UofC2011_OriT_oriV.png|frame|center|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> | ||
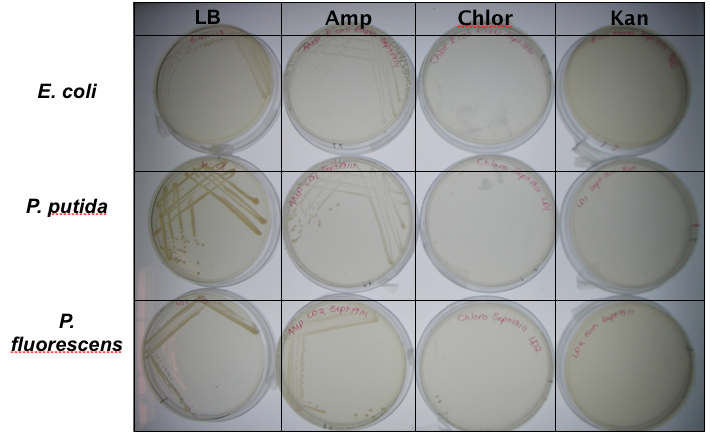
<p>In order to test our constructs were working, we needed a marker that we select for. We performed a growth assay on our <i>Pseudomonas</i> strains, verifying if they can grow on different antibiotics. <i>Pseudomonas</i> was streaked on LB agar plates containing no antibiotics, tetracycline (25 mg/ml), chloramphenicol (35 mg/mL), kanamycin (50 mg/mL), ampicillin (100 mg/mL) and gentamyacin (50 mg/mL) (figure x). Growth was observed on LB alone as well as on ampicillin, indicating that <i>Pseudomonas</i>, as expected, is naturally resistant to ampicillin. The lack of growth on the other antibiotics, however, indicated that resistance markers for these could possibly be used to test out the functionality of our constructs.</p> | <p>In order to test our constructs were working, we needed a marker that we select for. We performed a growth assay on our <i>Pseudomonas</i> strains, verifying if they can grow on different antibiotics. <i>Pseudomonas</i> was streaked on LB agar plates containing no antibiotics, tetracycline (25 mg/ml), chloramphenicol (35 mg/mL), kanamycin (50 mg/mL), ampicillin (100 mg/mL) and gentamyacin (50 mg/mL) (figure x). Growth was observed on LB alone as well as on ampicillin, indicating that <i>Pseudomonas</i>, as expected, is naturally resistant to ampicillin. The lack of growth on the other antibiotics, however, indicated that resistance markers for these could possibly be used to test out the functionality of our constructs.</p> | ||
| - | </html>[[Image:UofC2011 Growth assay data.png|frame|center|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> | + | </html>[[Image:UofC2011 Growth assay data.png|frame|center|150pix|''Pseudomonas'' (left), microalgae species ''Dunaliella tertiolecta'' (right).]]<html> |
<p>Based on this, we moved our constructs into tetracycline and kanamycin resistance vectors for testing. Colonies of <i>E.Coli</i> containing our plasmids were used to innocluate 5 mL LB cultures with kanamycin-50 and tetracycline-25. 100 μL of our two <i>Pseudomonas</i> strains were added and cultures were shaken in a 26°C incubator. OD readings of cultures were taken to standardize cell counts and cultures were then plated on MacConkey agar plates. MacConkey agar, which is a reddish-purple color, is able to distinguish between bacteria that can and cannot metabolize lactose via a color change. Bacteria that do not metabolize lactose, such as <i>Pseudomonas</i>, use peptone instead and reduce ammonia, which causes a yellow color change of the plates due to an increase in pH. <i>E. coli</i>, on the other hand, do metabolize lactose, and therefore no color change is seen.</p> | <p>Based on this, we moved our constructs into tetracycline and kanamycin resistance vectors for testing. Colonies of <i>E.Coli</i> containing our plasmids were used to innocluate 5 mL LB cultures with kanamycin-50 and tetracycline-25. 100 μL of our two <i>Pseudomonas</i> strains were added and cultures were shaken in a 26°C incubator. OD readings of cultures were taken to standardize cell counts and cultures were then plated on MacConkey agar plates. MacConkey agar, which is a reddish-purple color, is able to distinguish between bacteria that can and cannot metabolize lactose via a color change. Bacteria that do not metabolize lactose, such as <i>Pseudomonas</i>, use peptone instead and reduce ammonia, which causes a yellow color change of the plates due to an increase in pH. <i>E. coli</i>, on the other hand, do metabolize lactose, and therefore no color change is seen.</p> | ||
</html> | </html> | ||
}} | }} | ||
Revision as of 02:37, 28 September 2011









A Chassis Using Native Tailings Pond Species

Chassis Selection
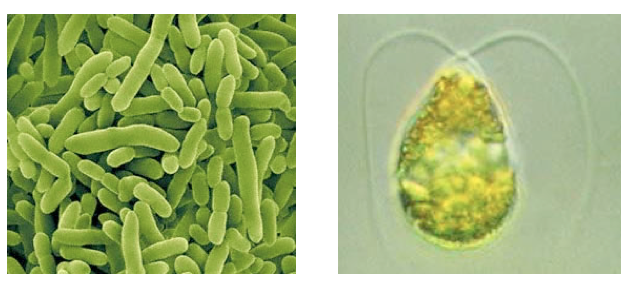
We identified early on in our project planning stages that we would eventually want to use Pseudomonas or microalgae as the chassis to house our system. The motivation for this is that not only are these organisms capable of naturally surviving a tailings ponds environment, but they may contain necessary regulatory elements required for a response to NA's. Because of this, taking a response element out of an organism and putting our system into another chassis, such as E. coli, may not behave as expected. Based on this chassis choice however, we realized that given their scarcity in iGEM, tools needed to establish work in these organisms for future iGEM teams.Microalgae Tools
In terms of microalgae, the first things we needed were protocols for various procedures. We wanted to eventually use our microagae strain to construct a promoter library out of nuclear and chloroplast DNA. As such, transformation as well as nuclear DNA and chloroplast extraction protocols were necessary. Unfortunately, our strain of microalgae, Dunaliella tertiolecta, has not been transformed very often in the literature, and as such we had to spend some time optimizing literature protocols. All microalgae protocols we used can be found in our protocol section.
Transformation
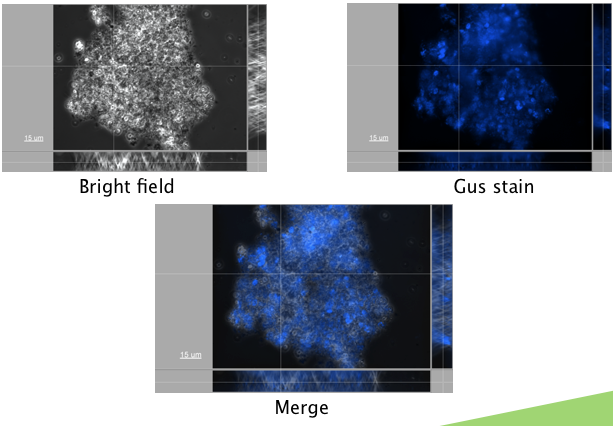
An appropriate reporter also needed to be found. A reporter construct containing GFP downstream of a Nopaline Synthase (NOS) promoter was first transformed. Autofluorescence levels were found to be too high, however, to detect fluorescence above the background, making this a poor choice for a reporter. A GUS reporter commonly used in plants, and available in the registry (BBa_K330002) was tried next. GUS (beta-glucuronidase) is an enzyme from E. coli, that is frequently used a s a plant reporter. A construct conatining the GUS reporter gene, downstream of the NOS reporter was successfully transformed (figure x.)
A Pseudomonas Conjugation System
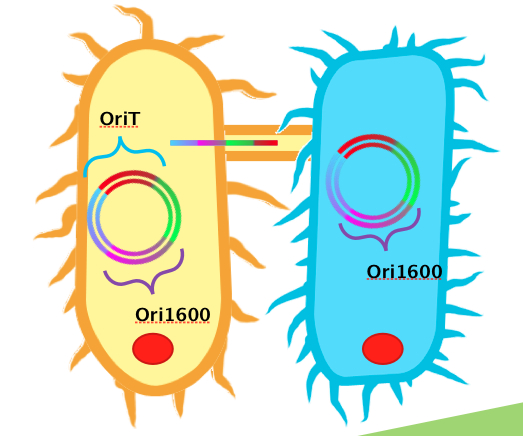
In terms of Pseudomonas, we realized that we would need a way to easily and cheaply transform Pseudomonas with high efficiency. To do this, we decided to design a conjugation system, capable of conjugating plasmids from E. coli to Pseudomonas. In order for conjugation processes to be possible from E. coli to Pseudomonas spp., E. coli requires plasmids containing all the necessary genes and genetic elements. In order for a plasmid to transfer by conjugation, it needs a sequence recognizable by relaxase and nickase called oriT, which indicates the origin of transfer. The plasmid to be transferred also requires replication origins for both E. coli (such as ColE1) and Pseudomonas spp. (such as ori1600). Ori1600, which we used in our system is a replication-stabilizing element for Pseudomonas spp., and requires ColE1 to function. In addition, the tra region contains all the genes necessary for the conjugation process (nickase, pili formation proteins, DNA polymerases etc.), and is either native to the E. coli strain on a F/fertility plasmid, or needs to be supplied in the form of a helper strain (e.g. pRK2013 contains a RK4 plasmid that supplies all the tra genes).
We obtained DNA for both oriT and ori1600, BioBricked them and have submitted them as BioBrick parts. We then constructed them together using BioBrick cloning, as a testing construct. Already present in the registry is an oriT part J01003, which was characterized by Heidelberg in 2008. This part was used for conjugation between strains of E. coli. Technically, this part could also be used for conjugation between Pseudomonas and E. coli. Based on the sequences however, we hypothesized that our oriT, which is considerably longer, may have additional regions to improve efficiency in conjugating to Pseudomonas. To test this, we built additional constructs using J01003 and our ori1600 part to compare to our oriT- ori1600 construct.
In order to test our constructs were working, we needed a marker that we select for. We performed a growth assay on our Pseudomonas strains, verifying if they can grow on different antibiotics. Pseudomonas was streaked on LB agar plates containing no antibiotics, tetracycline (25 mg/ml), chloramphenicol (35 mg/mL), kanamycin (50 mg/mL), ampicillin (100 mg/mL) and gentamyacin (50 mg/mL) (figure x). Growth was observed on LB alone as well as on ampicillin, indicating that Pseudomonas, as expected, is naturally resistant to ampicillin. The lack of growth on the other antibiotics, however, indicated that resistance markers for these could possibly be used to test out the functionality of our constructs.
Based on this, we moved our constructs into tetracycline and kanamycin resistance vectors for testing. Colonies of E.Coli containing our plasmids were used to innocluate 5 mL LB cultures with kanamycin-50 and tetracycline-25. 100 μL of our two Pseudomonas strains were added and cultures were shaken in a 26°C incubator. OD readings of cultures were taken to standardize cell counts and cultures were then plated on MacConkey agar plates. MacConkey agar, which is a reddish-purple color, is able to distinguish between bacteria that can and cannot metabolize lactose via a color change. Bacteria that do not metabolize lactose, such as Pseudomonas, use peptone instead and reduce ammonia, which causes a yellow color change of the plates due to an increase in pH. E. coli, on the other hand, do metabolize lactose, and therefore no color change is seen.

 "
"