RecA Project
From 2011.igem.org
(→RecA Co-Protease) |
(→RecA Co-Protease) |
||
| Line 42: | Line 42: | ||
[[File:RecATest2.png|center|]] | [[File:RecATest2.png|center|]] | ||
| + | ==Method== | ||
| + | <p>The RecA group focused its resources on mutating RecA1 as described above. To perform these mutations, site directed mutagenesis was performed on the RecA1 plasmid. The single primer method was used in an attempt to perform all five mutations at one time. The protocol for this procedure can be found on the [[Protocols| protocols page]], and was taken from the Knight Lab openwetware page. After a summer of failure with this method (only one mutation in three months), the Penn State team switched to the double primer method using a protocol based off of the protocol from the Richard Lab openwetware page. The modified protocol can be found on the [[Protocols| protocols page]].</p> | ||
| - | + | <p>When using the double primer method of site directed mutagenesis, only one base pair can be mutated at one time. Thus, the Penn State team redefined their goal regarding RecA1 mutagenesis in order to expand their data set. By mutating each site at one time, the Penn State team can use the RecA test circuit to determine the effectiveness of each mutation as well as each combination of mutations. The EcoRI and PstI sites had to be mutated in every RecA plasmid in order for BioBrick assembly, which was needed to put the RecA test circuit together, to work correctly. Once these sites were mutated, the plasmid containing these two mutations was then used to mutate the Lys, Arg, and RecA activation sites individually. Once these sites were mutated, the plasmids were used to mutate a different site that did not already exist in the plasmid. Finally, the final site was mutated, resulting in a plasmid that contained all five mutations in RecA1 to form RecA. This left seven total mutated RecA1 plasmids that could be tested using the RecA test circuit. The Penn State mutagenesis plan can be summarized by the below picture.</p> | |
Revision as of 05:56, 27 September 2011

RecA Project Sensor Project Reporter Project
RecA Co-Protease
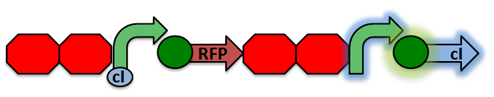
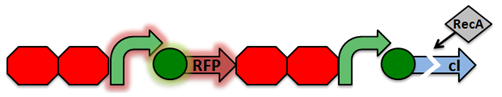
The normal laboratory strain of E. coli, DH10B, contains an inactivated form of RecA known as RecA1. Therefore, a series of mutations must occur in order to restore its proteatic function and reduce recombination.
Activation of RecA
- Point Mutation at b.p 720: A ⇒ G
Removal of Recombinase Activity
- Arginine 243 ⇒ Glutamine
- Lysine 286 ⇒ Asparagine
In order to use the standard bio-bricking techniques, we also had to remove these naturally occurring enzymatic restriction sites
- PST1 Site [CTGCAG]
- CAG ⇒ CTG (Codon usage bias decreases: .69 to .31)
- EcoR1 Site [GAATTC]
- TTC ⇒ TTT (Codon usage bias increases: .49 to .51)
Activated RecA then cleaves the cI repressor, causing transcription of RFP. However, if RecA exhibits recombinant activity, then the homologous double terminator regions will recombine. This causes the deletion of the repressible promoter and RFP.
Method
The RecA group focused its resources on mutating RecA1 as described above. To perform these mutations, site directed mutagenesis was performed on the RecA1 plasmid. The single primer method was used in an attempt to perform all five mutations at one time. The protocol for this procedure can be found on the protocols page, and was taken from the Knight Lab openwetware page. After a summer of failure with this method (only one mutation in three months), the Penn State team switched to the double primer method using a protocol based off of the protocol from the Richard Lab openwetware page. The modified protocol can be found on the protocols page.
When using the double primer method of site directed mutagenesis, only one base pair can be mutated at one time. Thus, the Penn State team redefined their goal regarding RecA1 mutagenesis in order to expand their data set. By mutating each site at one time, the Penn State team can use the RecA test circuit to determine the effectiveness of each mutation as well as each combination of mutations. The EcoRI and PstI sites had to be mutated in every RecA plasmid in order for BioBrick assembly, which was needed to put the RecA test circuit together, to work correctly. Once these sites were mutated, the plasmid containing these two mutations was then used to mutate the Lys, Arg, and RecA activation sites individually. Once these sites were mutated, the plasmids were used to mutate a different site that did not already exist in the plasmid. Finally, the final site was mutated, resulting in a plasmid that contained all five mutations in RecA1 to form RecA. This left seven total mutated RecA1 plasmids that could be tested using the RecA test circuit. The Penn State mutagenesis plan can be summarized by the below picture.
 "
"