Team:Harvard/Results/MAGE
From 2011.igem.org
| Line 1: | Line 1: | ||
| - | {{:Team:Harvard/Template: | + | {{:Team:Harvard/Template:ResultsBar}} |
{{:Team:Harvard/Template:ResultsGrayBar}} | {{:Team:Harvard/Template:ResultsGrayBar}} | ||
<!-- | <!-- | ||
Revision as of 05:10, 26 September 2011
For a detailed explanation of how MAGE works, check out our MAGE technology page.
For a step-by-step procedure, see Protocols.
Building the selection strain: MAGE
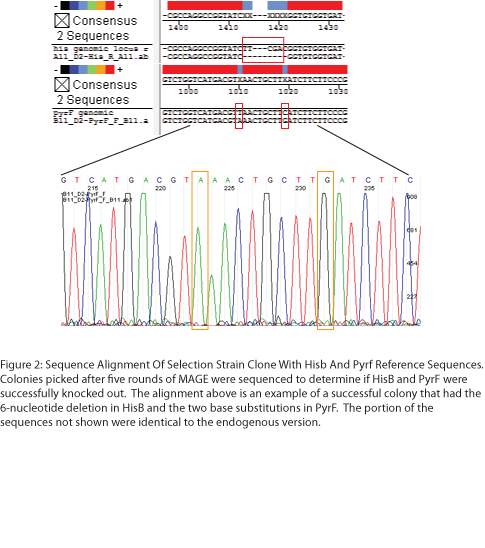
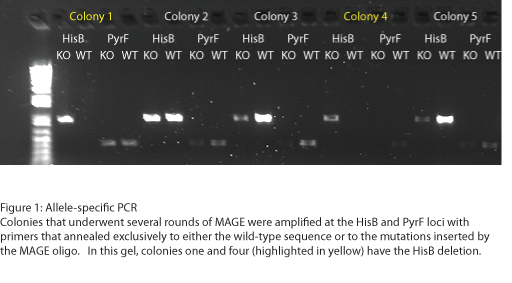
To test the zinc fingers constructed by chip synthesis, we used a one-hybrid system tying zinc finger binding to cell survival (Meng et al, 2005). Under the control of a zinc finger binding site were two yeast genes: His3, a positive metabolic selector, and URA3, a negative selector to eliminate leaky promoters. To create a selection strain for this system, we accordingly knocked out the endogenous E. coli homologs (HisB and PyrF, respectively) using multiplex automated genome engineering (MAGE). The PyrF MAGE oligo inserted two early in-frame stop codons by substituting 8T-->A and 16C-->G, while the HisB oligo deleted 6 in-frame nucleotides (694-699) coding for VE. 5 rounds of MAGE were performed in an EcNR2 E. coli line using both oligos simultaneously (see Protocols for exact procedure) and the resulting colonies underwent allele-specific PCR (Figure 1) and were sequenced at the HisB and PyrF loci (Figure 2). We chose a colony that contained both the HisB deletion and the PyrF substitutions as the basis of our selection strain.
References:
Xiangdong Meng, Michael H Brodsky, Scot A Wolfe. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. (2005) Nature Biotechnology, 23(8): 988-994.
 "
"