Team:Kyoto/Lab Work
From 2011.igem.org
(→Week7: Monday 12th September - Sunday 18th September) |
|||
| Line 297: | Line 297: | ||
:{|class="wikitable" | :{|class="wikitable" | ||
|+Reaction mixture | |+Reaction mixture | ||
| - | ! | + | !Component||Volume(μl) |
| - | + | ||
|- | |- | ||
|2x Buffer||25 | |2x Buffer||25 | ||
| Line 309: | Line 308: | ||
|- | |- | ||
|Template||X | |Template||X | ||
| + | |- | ||
| + | |KOD FX||1 | ||
|- | |- | ||
|ddH<sub>2</sub>O||up to 50 | |ddH<sub>2</sub>O||up to 50 | ||
| Line 316: | Line 317: | ||
|Predenature||94C||2m|| | |Predenature||94C||2m|| | ||
|- | |- | ||
| - | |Denature||98C||10s||rowspan="3"| | + | |Denature||98C||10s||rowspan="3"|30cycles |
|- | |- | ||
|Annealing||56C||30s | |Annealing||56C||30s | ||
| Line 331: | Line 332: | ||
'''Saturday'''<br/> | '''Saturday'''<br/> | ||
| + | |||
| + | Digestion(Kajita) | ||
| + | Retry of PCR amplification of SAM-P20 and ChiA. | ||
| + | |||
| + | :We amplified SAM-P20 by colony direct PCR and ChiA by PCR using the PCR product that performed yesterday. | ||
| + | |||
| + | :Reaction mixture: The same components and volume as before. | ||
| + | |||
| + | :PCR condition: The same PCR condition as before. | ||
| + | |||
| + | |||
| + | :PCR for SAM-P20 | ||
| + | |||
| + | :We prepared two kinds of templates: | ||
| + | #Picked a colony, suspended in 50ul of TE buffer (pH8.0) and incubated 1min at 95 degree. Added 5ul to the reaction mixture. | ||
| + | #Picked a colony and dipped in the reaction mixture. | ||
| + | |||
| + | :PCR for ChiA | ||
| + | :1μl of PCR product was added to the reaction mixture as template. | ||
| + | |||
'''Sunday'''<br/> | '''Sunday'''<br/> | ||
Revision as of 16:54, 22 September 2011
Lab Work
Week1: Monday 1st - Sunday 7th August
Monday
行った実験名:
使った試薬名、容量:
用いた機械:
行った人:
Tuesday
Wednesday
Thursday
Friday
Saturday
Sunday
Week2: Monday 8th - Sunday 14th August
Monday
Tuesday
Wednesday
Thursday
Friday
Saturday
Sunday
Week3: Monday 15th - Sunday 21th August
Monday
Tuesday
Wednesday
Thursday
Nutritional Signal(Sugiura,Shimosaka & Okumura)
PCR Amplification of glnL and glnG from gDNA of E.coli
--Primers--
glnL
Left primer: tctagaggagactgctttatggcaac
Right primer: actagtaggaactatcgtcatcgactac
glnG
Left primer: tctagaggtgacgtttatgcaacga
Right primer: actagtacacacaagctgtgaatcactc
annealing temperature was 55 degrees.
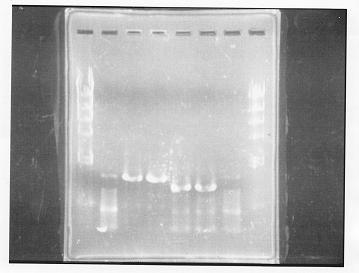
lane 1,2 &7,8: DNA ladder(λDNA digested Hindlll,100bp), lane 3,4: glnG1,glnG2, lane 5,6: glnL1,glnL2
After purification, the concentration of DNA are
glnG1: 127.8ng/ul
glnG2: 118.1ng/ul
glnL1: 137.4ng/ul
glnL2: 124.2ng/ul
Friday
Nutritional Signal(Shimosaka)
Restriction of glnG1 and glnG2
Cut them with Xbal and Spel for 2 hours at 37 degrees.
Then, gel extraction of digested.
Saturday
Sunday
Week4: Monday 22th - Sunday 28th August
Monday
Tuesday
Wednesday
Thursday
Luminescence(Kusaba, Terada, Hara):ハエの走行性実験① ♂、紫外線×2回、緑×2回 ♀、紫外線×2回、緑×2回
Friday
Luminescence(Kusaba):ハエの走行性実験① ♂、赤外線×2回 ♀、赤外線×2回
Nutritional Signal(Hashiya):
Transformation of bellow parts.
4-17M:BBa_K325909(lux operon)
1-12M:BBa_E0240
2-17F:BBa_120260(low copy vector)
PCR amplification of
Saturday
Luminescence(Kusaba):ハエの走行性実験① ♂、赤×2回、青×2回 ♀、赤×2回、青×2回
Sunday
Week5: Monday 29th August - Sunday 4th September
Monday
Tuesday
Nutritional Signal(Hashiya)
・PCR amplification of glnL and glnG from PCR products,glnL1 and glnG1.
--Primers--
glnL
Left primer: ggaattcgcggccgcttctagaggagactgctttatggcaac
Right primer: ggactagtaggaactatcgtcatcgactac
glnG
Left primer: ggaattcgcggccgcttctagaggtgacgtttatgcaacga
Right primer: ggactagtacacacaagctgtgaatcactc
annealing temperature was 55 degrees.
・PCR amplification of glnL+G and rpoN from gDNA of E.coli.
--Primers--
glnL+G
Left primer: ggaattcgcggccgcttctagaggagactgctttatggcaac
Right primer: ggactagtacacacaagctgtgaatcactc
rpoN
Left primer: ggaattcgcggccgcttctagaggttctgaacatgaagcaa
Right primer: ggactagtatccttatcggttgggtca
annealing temperature was 56 degrees.
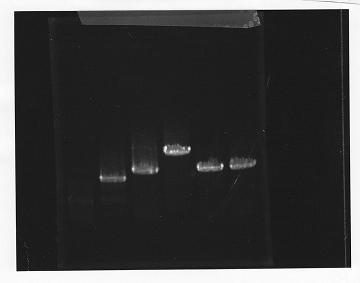
lane1: 100bp DNA ladder, lane2:glnL, lane3:glnG, lane4:glnG+L, lane5:rpoN from gDNA, lane6:rpoN from ASKA clone
After purification, the concentration of DNA are
glnL: 122.3 ng/ul
glnG: 64.7 ng/ul
glnL+G: 106.7 ng/ul
rpoN from gDNA: 111.4 ng/ul
Wednesday
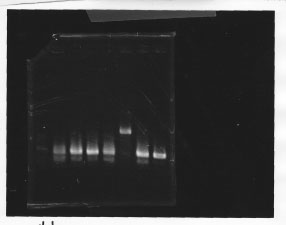
Nutritional Signal(Hashiya)
・Screening PCR of σ54 promoter + pSB1A3
We cultured σ54 promoter5
Friday
Nutritional Signal(Hashiya)
・Restriction of σ54 promoter5,glnL, glnG, glnL+G and rpoN
Cut them with EcoRl and Spel
After purification, the concentration of DNA were
σ54 promoter5: 23.6 ng/ul
glnL: 28.1 ng/ul
glnG: 26.3 ng/ul
glnL+G: 15.3 ng/ul
rpoN: 20.8 ng/ul
・Ligation reaction
Ligated glnL, glnG and rpoN to pSB1K3.
Thursday
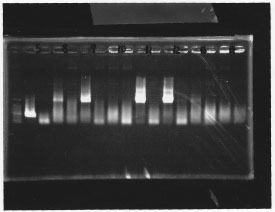
Nutritional Signal(Hashiya)
・Screening PCR of glnL, glnG, glnL+G and rpoN
glnL
glnG

glnL+G & rpoN
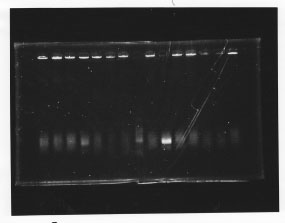
We cultured glnL5, glnG4
Saturday
Nutritional Signal(Hashiya)
・Mini prep of glnL5 and glnG4
glnL5: 43.9 ng/ul
glnG4: 38.1 ng/ul
Predation(Hashiya)
・PCR amplification of glmS
--Primers--
left primer:ggaattcgcggccgcttctagagcaggttgaccgacaacgata
right primer:ggactagtacgcagggcatccatttat
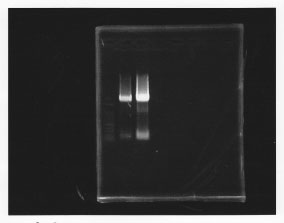
lane1: 100bp DNA ladder, lane2: glmS from gDNA, lane3: glmS from ASKA clone
・TA cloning of glmS
Ligated glmS from gDNA to pTA vector.
Sunday
Nutritional Signal(Hashiya)
・Screening PCR of rpoN

・Transformation of bellow parts
1-23L: BBa_B0015 (double terminator)
1-18E: BBa_J23101 (constitutive promoter)
1-18C: BBa_J23100 (constitutive promoter)
Week6: Monday 5th September - Sunday 11th September
Monday
Nutritional Signal(Hashiya)
・Screening PCR of glnL+G+double terminator
Tuesday
Luminescence(Kusaba, Hara):ハエの走行性実験②(②は改良版) ♂、緑×2回 ♀、青×1回
Wednesday
Luminescence(Hara):ハエの走行性実験② ♂、青×2回 ♀、緑×2回、青×1回
Thursday
Luminescence(Hara):ハエの走行性実験② ♂、紫外線×3回 ♀、紫外線×3回
Friday
Saturday
Luminescence(Kusaba, Hara):ハエの走行性実験② ♂、青×2回、赤×2回 ♀、青×2回、赤×2回
Sunday
Luminescence(Kusaba, Hara):ハエの走行性実験② ♂、紫外線×2回、
Week7: Monday 12th September - Sunday 18th September
Monday
Luminescence:大腸菌の形質転換(Hashiya) ハエの走行性実験②(Kusaba, Hara)
Tuesday
Luminescence:大腸菌はじめて光る。しかし光量は少ない。
Wednesday
Thursday
Friday
Digestion(Kajita)
PCR amplification of SAM-P20 and ChiA
- We performed colony direct PCRs from a S. albogriseolus colony and a S. avermitilis colony.
Reaction mixture Component Volume(μl) 2x Buffer 25 2mM dNTPs 10 Primer 1 1.5 Primer 2 1.5 Template X KOD FX 1 ddH2O up to 50
PCR condition Predenature 94C 2m Denature 98C 10s 30cycles Annealing 56C 30s Extension 68C 1m30s
- We prepared two kinds of templates:
- Picked a colony, suspended in 50ul of water and incubated 1min at 95 degree. Added 1ul to the reaction mixture.
- Picked a colony and dipped in the reaction mixture.
- Gel electrophoresis indicated that the PCR amplifications were successful for a sample, ChiA, but it was a faint band. We decided to retry direct PCR of SAM-P20 and PCR of ChiA.
Saturday
Digestion(Kajita) Retry of PCR amplification of SAM-P20 and ChiA.
- We amplified SAM-P20 by colony direct PCR and ChiA by PCR using the PCR product that performed yesterday.
- Reaction mixture: The same components and volume as before.
- PCR condition: The same PCR condition as before.
- PCR for SAM-P20
- We prepared two kinds of templates:
- Picked a colony, suspended in 50ul of TE buffer (pH8.0) and incubated 1min at 95 degree. Added 5ul to the reaction mixture.
- Picked a colony and dipped in the reaction mixture.
- PCR for ChiA
- 1μl of PCR product was added to the reaction mixture as template.
Sunday
Week8: Monday 19th September - Sunday 25th September
Monday
Tuesday
Wednesday
Thursday
Friday
Saturday
Sunday
Week9: Monday 26th September - Sunday 2nd October
Monday
Tuesday
Wednesday
Thursday
Friday
Saturday
Sunday
Protocol
Medium for drosophila
[http://www.biol.se.tmu.ac.jp/fly/www/standard-medium.html Medium for drosophila] Materials Methods - water : 500mL
- dry yeast : 20g
- corn flour : 45g
- glucose : 50g
- agarose : 3.5~5g
- propionic acid : 1.5mL
- 10% p-hydroxybenzoate in 70% Eternol : 5g
- Stir dry yeast and agarose with about two-thirds of water. Then, autoclave it.
- Stir corn flour and glucose with the remaining water.
- Stir 1 and 2, then autoclave it again.
- after autoclave, add propionic acid and 10% p-hydroxybenzoate in 70% Eternol into it.■
 "
"






